ARFS vs Boruta and BorutaShap#
comparison with Leshy, which is BorutaPy implementation with:
categorical features handling
plot method
catboost and lightGBM handling
SHAP and permutation importance
sample weight
The implementation is however quite close to the BorutaPy one. A PR has been opened on the official BorutaPy repo.
[1]:
# from IPython.core.display import display, HTML
# display(HTML("<style>.container { width:95% !important; }</style>"))
import numpy as np
import pandas as pd
import matplotlib as mpl
import matplotlib.pyplot as plt
import gc
import shap
from boruta import BorutaPy as bp
from sklearn.datasets import fetch_openml
from sklearn.inspection import permutation_importance
from sklearn.pipeline import Pipeline
from sklearn.datasets import fetch_openml
from sklearn.inspection import permutation_importance
from sklearn.base import clone
from sklearn.ensemble import RandomForestRegressor, RandomForestClassifier
from sys import getsizeof, path
from boruta import BorutaPy
import arfs
import arfs.feature_selection as arfsfs
import arfs.feature_selection.allrelevant as arfsgroot
from arfs.utils import LightForestClassifier, LightForestRegressor
from arfs.benchmark import highlight_tick, compare_varimp, sklearn_pimp_bench
from arfs.utils import load_data
# plt.style.use('fivethirtyeight')
rng = np.random.RandomState(seed=42)
# import warnings
# warnings.filterwarnings('ignore')
Using `tqdm.autonotebook.tqdm` in notebook mode. Use `tqdm.tqdm` instead to force console mode (e.g. in jupyter console)
[2]:
%matplotlib inline
[3]:
gc.enable()
gc.collect()
[3]:
0
Comparison#
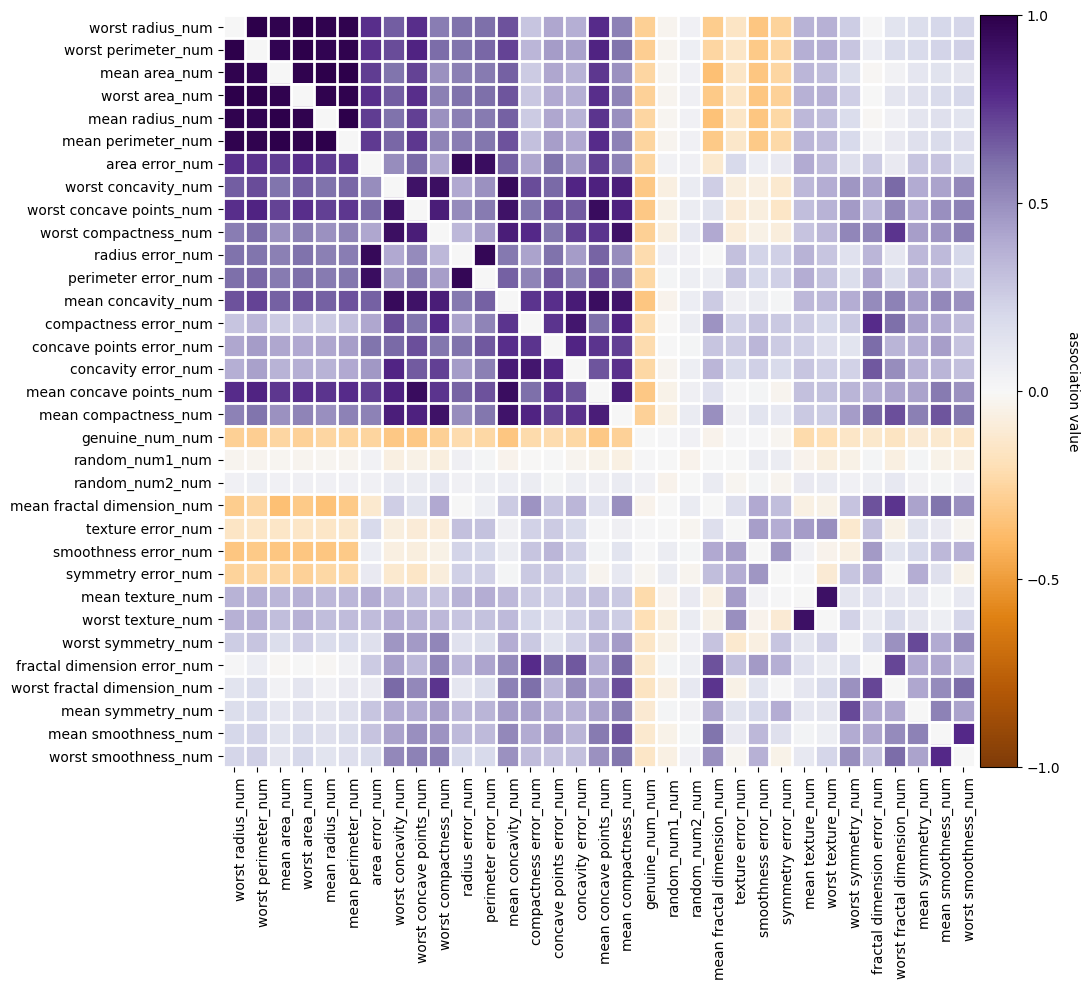
I’ll just remove the collinear predictors since they are actually harmful for the ARFS, see the Collinearity notebook
[4]:
cancer = load_data(name="cancer")
X, y = cancer.data, cancer.target
# basic feature selection
basic_fs_pipeline = Pipeline(
[
("missing", arfsfs.MissingValueThreshold(threshold=0.05)),
("unique", arfsfs.UniqueValuesThreshold(threshold=1)),
("cardinality", arfsfs.CardinalityThreshold(threshold=1000)),
("collinearity", arfsfs.CollinearityThreshold(threshold=0.75)),
]
)
X_filtered = basic_fs_pipeline.fit_transform(
X=X, y=y
) # , collinearity__sample_weight=w,
X_filtered.head()
[4]:
| mean texture | mean area | texture error | smoothness error | symmetry error | worst smoothness | random_num1 | random_num2 | genuine_num | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | 10.38 | 1001.0 | 0.9053 | 0.006399 | 0.03003 | 0.1622 | 0.496714 | 0 | -0.249340 |
| 1 | 17.77 | 1326.0 | 0.7339 | 0.005225 | 0.01389 | 0.1238 | -0.138264 | 1 | -0.044410 |
| 2 | 21.25 | 1203.0 | 0.7869 | 0.006150 | 0.02250 | 0.1444 | 0.647689 | 3 | 0.128395 |
| 3 | 20.38 | 386.1 | 1.1560 | 0.009110 | 0.05963 | 0.2098 | 1.523030 | 0 | -0.079921 |
| 4 | 14.34 | 1297.0 | 0.7813 | 0.011490 | 0.01756 | 0.1374 | -0.234153 | 0 | -0.094302 |
[5]:
f = basic_fs_pipeline.named_steps["collinearity"].plot_association()
BorutaPy#
Boruta, in its “official” implementation uses gain/gini feature importance (which is known to be biased). Let’s see what are the results on this data set
Unfortunately, Boruta does not work with newer version of numpy and python. However, if you have an older version of numpy, yuo can run this comparison, it should returns the same results.
[6]:
%%time
# define random forest classifier, with utilising all cores and
# sampling in proportion to y labels
rf = RandomForestClassifier(n_jobs=-1, class_weight="balanced", max_depth=5)
# define Boruta feature selection method
bp_feat_selector = BorutaPy(rf, n_estimators="auto", verbose=1, random_state=1)
# find all relevant features - 5 features should be selected
bp_feat_selector.fit(X_filtered.values, y.values)
# check selected features - first 5 features are selected
print("\n")
print(list(X_filtered.columns[bp_feat_selector.support_]))
---------------------------------------------------------------------------
AttributeError Traceback (most recent call last)
File <timed exec>:9
File ~/mambaforge-pypy3/envs/arfs/lib/python3.10/site-packages/boruta/boruta_py.py:201, in BorutaPy.fit(self, X, y)
188 def fit(self, X, y):
189 """
190 Fits the Boruta feature selection with the provided estimator.
191
(...)
198 The target values.
199 """
--> 201 return self._fit(X, y)
File ~/mambaforge-pypy3/envs/arfs/lib/python3.10/site-packages/boruta/boruta_py.py:260, in BorutaPy._fit(self, X, y)
255 _iter = 1
256 # holds the decision about each feature:
257 # 0 - default state = tentative in original code
258 # 1 - accepted in original code
259 # -1 - rejected in original code
--> 260 dec_reg = np.zeros(n_feat, dtype=np.int)
261 # counts how many times a given feature was more important than
262 # the best of the shadow features
263 hit_reg = np.zeros(n_feat, dtype=np.int)
File ~/mambaforge-pypy3/envs/arfs/lib/python3.10/site-packages/numpy/__init__.py:305, in __getattr__(attr)
300 warnings.warn(
301 f"In the future `np.{attr}` will be defined as the "
302 "corresponding NumPy scalar.", FutureWarning, stacklevel=2)
304 if attr in __former_attrs__:
--> 305 raise AttributeError(__former_attrs__[attr])
307 # Importing Tester requires importing all of UnitTest which is not a
308 # cheap import Since it is mainly used in test suits, we lazy import it
309 # here to save on the order of 10 ms of import time for most users
310 #
311 # The previous way Tester was imported also had a side effect of adding
312 # the full `numpy.testing` namespace
313 if attr == 'testing':
AttributeError: module 'numpy' has no attribute 'int'.
`np.int` was a deprecated alias for the builtin `int`. To avoid this error in existing code, use `int` by itself. Doing this will not modify any behavior and is safe. When replacing `np.int`, you may wish to use e.g. `np.int64` or `np.int32` to specify the precision. If you wish to review your current use, check the release note link for additional information.
The aliases was originally deprecated in NumPy 1.20; for more details and guidance see the original release note at:
https://numpy.org/devdocs/release/1.20.0-notes.html#deprecations
Leshy#
Let’s compare to the official python implementation, using the same setting and the gini/gain feature importance. We should have the same results (btw, you can check the unit tests, BorutaPy is used as baseline).
[7]:
%%time
# Leshy, all the predictors, no-preprocessing
model = clone(rf)
leshy_feat_selector = arfsgroot.Leshy(
rf,
n_estimators="auto",
verbose=1,
max_iter=100,
random_state=1,
importance="native",
)
leshy_feat_selector.fit(X_filtered, y, sample_weight=None)
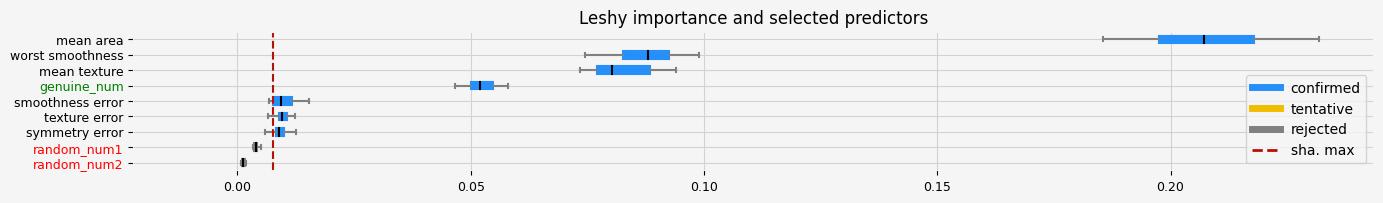
print(f"The selected features: {leshy_feat_selector.get_feature_names_out()}")
print(f"The agnostic ranking: {leshy_feat_selector.ranking_}")
print(f"The naive ranking: {leshy_feat_selector.ranking_absolutes_}")
fig = leshy_feat_selector.plot_importance(n_feat_per_inch=5)
# highlight synthetic random variable
fig = highlight_tick(figure=fig, str_match="random")
fig = highlight_tick(figure=fig, str_match="genuine", color="green")
plt.show()
fasttreeshap is not installed. Fallback to shap.
Leshy finished running using shap var. imp.
Iteration: 1 / 100
Confirmed: 7
Tentative: 0
Rejected: 2
All relevant predictors selected in 00:00:04.00
The selected features: ['mean texture' 'mean area' 'texture error' 'smoothness error'
'symmetry error' 'worst smoothness' 'genuine_num']
The agnostic ranking: [1 1 1 1 1 1 2 3 1]
The naive ranking: ['mean area', 'worst smoothness', 'mean texture', 'genuine_num', 'smoothness error', 'texture error', 'symmetry error', 'random_num1', 'random_num2']
CPU times: user 5.25 s, sys: 537 ms, total: 5.79 s
Wall time: 4.5 s
Same Results?#
[8]:
def check_list_equal(L1, L2):
return len(L1) == len(L2) and sorted(L1) == sorted(L2)
[9]:
check_list_equal(
leshy_feat_selector.get_feature_names_out(),
list(X_filtered.columns[bp_feat_selector.support_]),
)
---------------------------------------------------------------------------
AttributeError Traceback (most recent call last)
Cell In[9], line 3
1 check_list_equal(
2 leshy_feat_selector.get_feature_names_out(),
----> 3 list(X_filtered.columns[bp_feat_selector.support_]),
4 )
AttributeError: 'BorutaPy' object has no attribute 'support_'
BorutaShap with native importance#
BorutaShap, is an alternative implementation (heavy re-writting and new material) of Boruta with Shap feature importance. Let’s see what are the results on this data set
[10]:
%%time
from BorutaShap import BorutaShap
from arfs.preprocessing import OrdinalEncoderPandas
# define random forest classifier, with utilising all cores and
# sampling in proportion to y labels
model = RandomForestClassifier(n_jobs=-1, class_weight="balanced", max_depth=5)
# define BorutaShap feature selection method (doesn't convert automatically cat feature)
X_encoded = OrdinalEncoderPandas().fit_transform(X=X_filtered)
bs_feat_selector = BorutaShap(
model=model, importance_measure="gini", classification=True
)
# find all relevant features - 5 features should be selected
bs_feat_selector.fit(X=X_encoded, y=y, n_trials=100, random_state=0)
# Returns Boxplot of features
bs_feat_selector.plot(X_size=12, figsize=(8, 6), y_scale="log", which_features="all")
---------------------------------------------------------------------------
ImportError Traceback (most recent call last)
File <timed exec>:1
File ~/mambaforge-pypy3/envs/arfs/lib/python3.10/site-packages/BorutaShap.py:2
1 from sklearn.ensemble import RandomForestClassifier, RandomForestRegressor, IsolationForest
----> 2 from sklearn.datasets import load_breast_cancer, load_boston
3 from statsmodels.stats.multitest import multipletests
4 from sklearn.model_selection import train_test_split
File ~/mambaforge-pypy3/envs/arfs/lib/python3.10/site-packages/sklearn/datasets/__init__.py:157, in __getattr__(name)
108 if name == "load_boston":
109 msg = textwrap.dedent("""
110 `load_boston` has been removed from scikit-learn since version 1.2.
111
(...)
155 <https://www.researchgate.net/publication/4974606_Hedonic_housing_prices_and_the_demand_for_clean_air>
156 """)
--> 157 raise ImportError(msg)
158 try:
159 return globals()[name]
ImportError:
`load_boston` has been removed from scikit-learn since version 1.2.
The Boston housing prices dataset has an ethical problem: as
investigated in [1], the authors of this dataset engineered a
non-invertible variable "B" assuming that racial self-segregation had a
positive impact on house prices [2]. Furthermore the goal of the
research that led to the creation of this dataset was to study the
impact of air quality but it did not give adequate demonstration of the
validity of this assumption.
The scikit-learn maintainers therefore strongly discourage the use of
this dataset unless the purpose of the code is to study and educate
about ethical issues in data science and machine learning.
In this special case, you can fetch the dataset from the original
source::
import pandas as pd
import numpy as np
data_url = "http://lib.stat.cmu.edu/datasets/boston"
raw_df = pd.read_csv(data_url, sep="\s+", skiprows=22, header=None)
data = np.hstack([raw_df.values[::2, :], raw_df.values[1::2, :2]])
target = raw_df.values[1::2, 2]
Alternative datasets include the California housing dataset and the
Ames housing dataset. You can load the datasets as follows::
from sklearn.datasets import fetch_california_housing
housing = fetch_california_housing()
for the California housing dataset and::
from sklearn.datasets import fetch_openml
housing = fetch_openml(name="house_prices", as_frame=True)
for the Ames housing dataset.
[1] M Carlisle.
"Racist data destruction?"
<https://medium.com/@docintangible/racist-data-destruction-113e3eff54a8>
[2] Harrison Jr, David, and Daniel L. Rubinfeld.
"Hedonic housing prices and the demand for clean air."
Journal of environmental economics and management 5.1 (1978): 81-102.
<https://www.researchgate.net/publication/4974606_Hedonic_housing_prices_and_the_demand_for_clean_air>
The boston dataset is imported but removed from scikit-learn, from the version >= 1.2.0.
The hasattr is called before fitting so feature_importances_ is not found. Let’s use the default, which is also a random forest
[11]:
%%time
from BorutaShap import BorutaShap
bs_feat_selector = BorutaShap(importance_measure="gini", classification=True)
# find all relevant features - 5 features should be selected
bs_feat_selector.fit(X=X_encoded, y=y, n_trials=100, random_state=0)
# Returns Boxplot of features
bs_feat_selector.plot(X_size=12, figsize=(12, 8), y_scale="log", which_features="all")
---------------------------------------------------------------------------
ImportError Traceback (most recent call last)
File <timed exec>:1
File ~/mambaforge-pypy3/envs/arfs/lib/python3.10/site-packages/BorutaShap.py:2
1 from sklearn.ensemble import RandomForestClassifier, RandomForestRegressor, IsolationForest
----> 2 from sklearn.datasets import load_breast_cancer, load_boston
3 from statsmodels.stats.multitest import multipletests
4 from sklearn.model_selection import train_test_split
File ~/mambaforge-pypy3/envs/arfs/lib/python3.10/site-packages/sklearn/datasets/__init__.py:157, in __getattr__(name)
108 if name == "load_boston":
109 msg = textwrap.dedent("""
110 `load_boston` has been removed from scikit-learn since version 1.2.
111
(...)
155 <https://www.researchgate.net/publication/4974606_Hedonic_housing_prices_and_the_demand_for_clean_air>
156 """)
--> 157 raise ImportError(msg)
158 try:
159 return globals()[name]
ImportError:
`load_boston` has been removed from scikit-learn since version 1.2.
The Boston housing prices dataset has an ethical problem: as
investigated in [1], the authors of this dataset engineered a
non-invertible variable "B" assuming that racial self-segregation had a
positive impact on house prices [2]. Furthermore the goal of the
research that led to the creation of this dataset was to study the
impact of air quality but it did not give adequate demonstration of the
validity of this assumption.
The scikit-learn maintainers therefore strongly discourage the use of
this dataset unless the purpose of the code is to study and educate
about ethical issues in data science and machine learning.
In this special case, you can fetch the dataset from the original
source::
import pandas as pd
import numpy as np
data_url = "http://lib.stat.cmu.edu/datasets/boston"
raw_df = pd.read_csv(data_url, sep="\s+", skiprows=22, header=None)
data = np.hstack([raw_df.values[::2, :], raw_df.values[1::2, :2]])
target = raw_df.values[1::2, 2]
Alternative datasets include the California housing dataset and the
Ames housing dataset. You can load the datasets as follows::
from sklearn.datasets import fetch_california_housing
housing = fetch_california_housing()
for the California housing dataset and::
from sklearn.datasets import fetch_openml
housing = fetch_openml(name="house_prices", as_frame=True)
for the Ames housing dataset.
[1] M Carlisle.
"Racist data destruction?"
<https://medium.com/@docintangible/racist-data-destruction-113e3eff54a8>
[2] Harrison Jr, David, and Daniel L. Rubinfeld.
"Hedonic housing prices and the demand for clean air."
Journal of environmental economics and management 5.1 (1978): 81-102.
<https://www.researchgate.net/publication/4974606_Hedonic_housing_prices_and_the_demand_for_clean_air>
Comparison with Boruta#
[ ]:
check_list_equal(
list(X_filtered.columns[bp_feat_selector.support_]), list(bs_feat_selector.accepted)
)