Preprocessing - Basic feature selection#
Before performing All Relevant Feature Selection, you might want to apply preprocessing or basic feature selection to remove columns with:
a lot of missing values
zero variance
high cardinality
highly correlated (and keep only one)
zero predictive power
low predictive power
Any of those steps can be ignored but I recommend at least removing highly correlated predictors if you don’t have a good reason for keeping them in the dataset.
[1]:
# from IPython.core.display import display, HTML
# display(HTML("<style>.container { width:95% !important; }</style>"))
[2]:
# Settings and libraries
from __future__ import print_function
import pandas as pd
import numpy as np
import seaborn as sns
from matplotlib import pyplot as plt
import gc
import arfs
import arfs.preprocessing as arfspp
import arfs.feature_selection as arfsfs
from arfs.utils import (
_make_corr_dataset_regression,
_make_corr_dataset_classification,
)
%matplotlib inline
[3]:
print(f"Run with ARFS {arfs.__version__}")
Run with ARFS 2.2.3
[4]:
arfsfs.__all__
[4]:
['BaseThresholdSelector',
'MissingValueThreshold',
'UniqueValuesThreshold',
'CardinalityThreshold',
'CollinearityThreshold',
'VariableImportance',
'make_fs_summary',
'Leshy',
'BoostAGroota',
'GrootCV',
'MinRedundancyMaxRelevance',
'LassoFeatureSelection']
Generating artificial data, inspired by the BorutaPy unit test
[5]:
X, y, w = _make_corr_dataset_regression()
data = X.copy()
data["target"] = y
[6]:
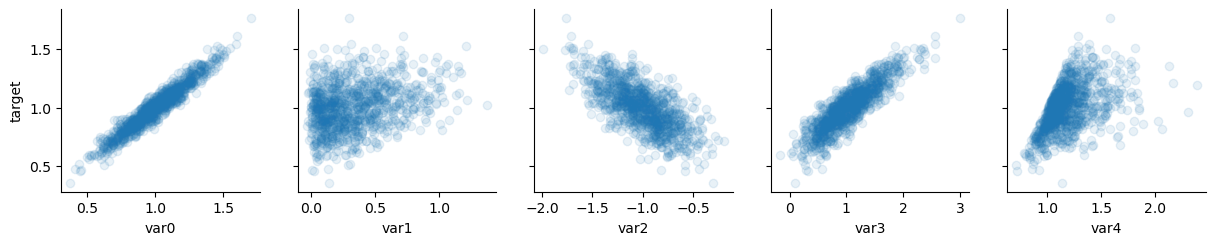
# significant regressors
x_vars = ["var0", "var1", "var2", "var3", "var4"]
y_vars = ["target"]
g = sns.PairGrid(data, x_vars=x_vars, y_vars=y_vars)
g.map(plt.scatter, alpha=0.1)
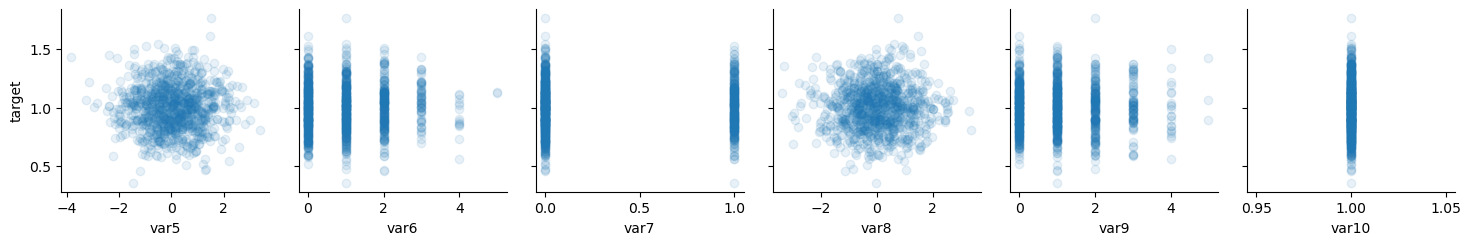
# noise
x_vars = ["var5", "var6", "var7", "var8", "var9", "var10"]
y_vars = ["target"]
g = sns.PairGrid(data, x_vars=x_vars, y_vars=y_vars)
g.map(plt.scatter, alpha=0.1)
plt.plot();
[7]:
X.head()
[7]:
| var0 | var1 | var2 | var3 | var4 | var5 | var6 | var7 | var8 | var9 | var10 | var11 | var12 | nice_guys | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1.016361 | 0.821664 | -1.184095 | 0.985738 | 1.050445 | -0.494190 | 2 | 0 | -0.361717 | 1 | 1.0 | 0 | 1.705980 | Bias |
| 1 | 0.929581 | 0.334013 | -1.063030 | 0.819273 | 1.016252 | 0.283845 | 1 | 0 | 0.178670 | 2 | 1.0 | 1 | NaN | Klaue |
| 2 | 1.095456 | 0.187234 | -1.488666 | 1.087443 | 1.140542 | 0.962503 | 2 | 0 | -3.375579 | 2 | 1.0 | 2 | NaN | Imaginedragons |
| 3 | 1.318165 | 0.994528 | -1.370624 | 1.592398 | 1.315021 | 1.165595 | 0 | 0 | -0.449650 | 2 | 1.0 | 3 | 2.929707 | MarkZ |
| 4 | 0.849496 | 0.184859 | -0.806604 | 0.865702 | 0.991916 | -0.058833 | 1 | 0 | 0.763903 | 1 | 1.0 | 4 | NaN | Thanos |
[8]:
X.dtypes
[8]:
var0 float64
var1 float64
var2 float64
var3 float64
var4 float64
var5 float64
var6 int64
var7 int64
var8 float64
var9 int64
var10 float64
var11 category
var12 float64
nice_guys object
dtype: object
[9]:
X.nunique()
[9]:
var0 1000
var1 1000
var2 1000
var3 1000
var4 1000
var5 1000
var6 6
var7 2
var8 1000
var9 6
var10 1
var11 500
var12 500
nice_guys 36
dtype: int64
[10]:
y = pd.Series(y)
y.name = "target"
Removing columns with many missing values#
[11]:
# X is the predictor DF (e.g: df[predictor_list]), at this stage you don't need to
# specify the target and weights (only for identifying zero and low importance)
# unsupervised learning, doesn't need a target
selector = arfsfs.MissingValueThreshold(0.05)
X_trans = selector.fit_transform(X)
print(f"The features going in the selector are : {selector.feature_names_in_}")
print(f"The support is : {selector.support_}")
print(f"The selected features are : {selector.get_feature_names_out()}")
The features going in the selector are : ['var0' 'var1' 'var2' 'var3' 'var4' 'var5' 'var6' 'var7' 'var8' 'var9'
'var10' 'var11' 'var12' 'nice_guys']
The support is : [ True True True True True True True True True True True True
False True]
The selected features are : ['var0' 'var1' 'var2' 'var3' 'var4' 'var5' 'var6' 'var7' 'var8' 'var9'
'var10' 'var11' 'nice_guys']
[12]:
X_trans.head()
[12]:
| var0 | var1 | var2 | var3 | var4 | var5 | var6 | var7 | var8 | var9 | var10 | var11 | nice_guys | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1.016361 | 0.821664 | -1.184095 | 0.985738 | 1.050445 | -0.494190 | 2 | 0 | -0.361717 | 1 | 1.0 | 0 | Bias |
| 1 | 0.929581 | 0.334013 | -1.063030 | 0.819273 | 1.016252 | 0.283845 | 1 | 0 | 0.178670 | 2 | 1.0 | 1 | Klaue |
| 2 | 1.095456 | 0.187234 | -1.488666 | 1.087443 | 1.140542 | 0.962503 | 2 | 0 | -3.375579 | 2 | 1.0 | 2 | Imaginedragons |
| 3 | 1.318165 | 0.994528 | -1.370624 | 1.592398 | 1.315021 | 1.165595 | 0 | 0 | -0.449650 | 2 | 1.0 | 3 | MarkZ |
| 4 | 0.849496 | 0.184859 | -0.806604 | 0.865702 | 0.991916 | -0.058833 | 1 | 0 | 0.763903 | 1 | 1.0 | 4 | Thanos |
[13]:
X.head()
[13]:
| var0 | var1 | var2 | var3 | var4 | var5 | var6 | var7 | var8 | var9 | var10 | var11 | var12 | nice_guys | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1.016361 | 0.821664 | -1.184095 | 0.985738 | 1.050445 | -0.494190 | 2 | 0 | -0.361717 | 1 | 1.0 | 0 | 1.705980 | Bias |
| 1 | 0.929581 | 0.334013 | -1.063030 | 0.819273 | 1.016252 | 0.283845 | 1 | 0 | 0.178670 | 2 | 1.0 | 1 | NaN | Klaue |
| 2 | 1.095456 | 0.187234 | -1.488666 | 1.087443 | 1.140542 | 0.962503 | 2 | 0 | -3.375579 | 2 | 1.0 | 2 | NaN | Imaginedragons |
| 3 | 1.318165 | 0.994528 | -1.370624 | 1.592398 | 1.315021 | 1.165595 | 0 | 0 | -0.449650 | 2 | 1.0 | 3 | 2.929707 | MarkZ |
| 4 | 0.849496 | 0.184859 | -0.806604 | 0.865702 | 0.991916 | -0.058833 | 1 | 0 | 0.763903 | 1 | 1.0 | 4 | NaN | Thanos |
Removing columns with zero variance#
[14]:
# single unique value(s) columns
# here rejecting columns with (or less than) 2 unique values
# unsupervised learning, doesn't need a target
selector = arfsfs.UniqueValuesThreshold(threshold=2)
X_trans = selector.fit_transform(X)
print(f"The features going in the selector are : {selector.feature_names_in_}")
print(f"The support is : {selector.support_}")
print(f"The selected features are : {selector.get_feature_names_out()}")
The features going in the selector are : ['var0' 'var1' 'var2' 'var3' 'var4' 'var5' 'var6' 'var7' 'var8' 'var9'
'var10' 'var11' 'var12' 'nice_guys']
The support is : [ True True True True True True True False True True False True
True True]
The selected features are : ['var0' 'var1' 'var2' 'var3' 'var4' 'var5' 'var6' 'var8' 'var9' 'var11'
'var12' 'nice_guys']
[15]:
X_trans.head()
[15]:
| var0 | var1 | var2 | var3 | var4 | var5 | var6 | var8 | var9 | var11 | var12 | nice_guys | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1.016361 | 0.821664 | -1.184095 | 0.985738 | 1.050445 | -0.494190 | 2 | -0.361717 | 1 | 0 | 1.705980 | Bias |
| 1 | 0.929581 | 0.334013 | -1.063030 | 0.819273 | 1.016252 | 0.283845 | 1 | 0.178670 | 2 | 1 | NaN | Klaue |
| 2 | 1.095456 | 0.187234 | -1.488666 | 1.087443 | 1.140542 | 0.962503 | 2 | -3.375579 | 2 | 2 | NaN | Imaginedragons |
| 3 | 1.318165 | 0.994528 | -1.370624 | 1.592398 | 1.315021 | 1.165595 | 0 | -0.449650 | 2 | 3 | 2.929707 | MarkZ |
| 4 | 0.849496 | 0.184859 | -0.806604 | 0.865702 | 0.991916 | -0.058833 | 1 | 0.763903 | 1 | 4 | NaN | Thanos |
[16]:
X.head()
[16]:
| var0 | var1 | var2 | var3 | var4 | var5 | var6 | var7 | var8 | var9 | var10 | var11 | var12 | nice_guys | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1.016361 | 0.821664 | -1.184095 | 0.985738 | 1.050445 | -0.494190 | 2 | 0 | -0.361717 | 1 | 1.0 | 0 | 1.705980 | Bias |
| 1 | 0.929581 | 0.334013 | -1.063030 | 0.819273 | 1.016252 | 0.283845 | 1 | 0 | 0.178670 | 2 | 1.0 | 1 | NaN | Klaue |
| 2 | 1.095456 | 0.187234 | -1.488666 | 1.087443 | 1.140542 | 0.962503 | 2 | 0 | -3.375579 | 2 | 1.0 | 2 | NaN | Imaginedragons |
| 3 | 1.318165 | 0.994528 | -1.370624 | 1.592398 | 1.315021 | 1.165595 | 0 | 0 | -0.449650 | 2 | 1.0 | 3 | 2.929707 | MarkZ |
| 4 | 0.849496 | 0.184859 | -0.806604 | 0.865702 | 0.991916 | -0.058833 | 1 | 0 | 0.763903 | 1 | 1.0 | 4 | NaN | Thanos |
Removing columns with high cardinality#
Large cardinality or many unique values are known to be favoured by tree-based models.
[17]:
# high cardinality for categoricals predictors
# unsupervised learning, doesn't need a target
selector = arfsfs.CardinalityThreshold(threshold=100)
X_trans = selector.fit_transform(X)
print(f"The features going in the selector are : {selector.feature_names_in_}")
print(f"The support is : {selector.support_}")
print(f"The selected features are : {selector.get_feature_names_out()}")
The features going in the selector are : ['var0' 'var1' 'var2' 'var3' 'var4' 'var5' 'var6' 'var7' 'var8' 'var9'
'var10' 'var11' 'var12' 'nice_guys']
The support is : [ True True True True True True True True True True True False
True True]
The selected features are : ['var0' 'var1' 'var2' 'var3' 'var4' 'var5' 'var6' 'var7' 'var8' 'var9'
'var10' 'var12' 'nice_guys']
[18]:
X_trans.head()
[18]:
| var0 | var1 | var2 | var3 | var4 | var5 | var6 | var7 | var8 | var9 | var10 | var12 | nice_guys | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1.016361 | 0.821664 | -1.184095 | 0.985738 | 1.050445 | -0.494190 | 2 | 0 | -0.361717 | 1 | 1.0 | 1.705980 | Bias |
| 1 | 0.929581 | 0.334013 | -1.063030 | 0.819273 | 1.016252 | 0.283845 | 1 | 0 | 0.178670 | 2 | 1.0 | NaN | Klaue |
| 2 | 1.095456 | 0.187234 | -1.488666 | 1.087443 | 1.140542 | 0.962503 | 2 | 0 | -3.375579 | 2 | 1.0 | NaN | Imaginedragons |
| 3 | 1.318165 | 0.994528 | -1.370624 | 1.592398 | 1.315021 | 1.165595 | 0 | 0 | -0.449650 | 2 | 1.0 | 2.929707 | MarkZ |
| 4 | 0.849496 | 0.184859 | -0.806604 | 0.865702 | 0.991916 | -0.058833 | 1 | 0 | 0.763903 | 1 | 1.0 | NaN | Thanos |
[19]:
X.head()
[19]:
| var0 | var1 | var2 | var3 | var4 | var5 | var6 | var7 | var8 | var9 | var10 | var11 | var12 | nice_guys | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1.016361 | 0.821664 | -1.184095 | 0.985738 | 1.050445 | -0.494190 | 2 | 0 | -0.361717 | 1 | 1.0 | 0 | 1.705980 | Bias |
| 1 | 0.929581 | 0.334013 | -1.063030 | 0.819273 | 1.016252 | 0.283845 | 1 | 0 | 0.178670 | 2 | 1.0 | 1 | NaN | Klaue |
| 2 | 1.095456 | 0.187234 | -1.488666 | 1.087443 | 1.140542 | 0.962503 | 2 | 0 | -3.375579 | 2 | 1.0 | 2 | NaN | Imaginedragons |
| 3 | 1.318165 | 0.994528 | -1.370624 | 1.592398 | 1.315021 | 1.165595 | 0 | 0 | -0.449650 | 2 | 1.0 | 3 | 2.929707 | MarkZ |
| 4 | 0.849496 | 0.184859 | -0.806604 | 0.865702 | 0.991916 | -0.058833 | 1 | 0 | 0.763903 | 1 | 1.0 | 4 | NaN | Thanos |
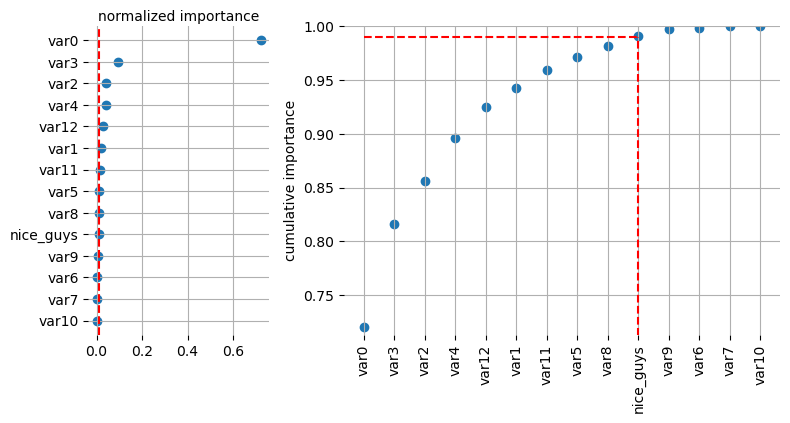
Removing columns with zero or low predictive power#
Identify the features with zero importance according to a gradient boosting machine. The lightgbm can be trained with early stopping using a utils set to prevent overfitting. The feature importances are averaged over n_iterations to reduce variance. Shapley values are used as feature importance for better results. If some predictors need to be encoded, integer encoding is chosen because OHE might lead to deep and unstable trees and lightGBM works great with integer encoding (see lightGBM
doc).
[25]:
lgb_kwargs = {"objective": "rmse", "zero_as_missing": False}
selector = arfsfs.VariableImportance(
verbose=2, threshold=0.99, lgb_kwargs=lgb_kwargs, fastshap=False
)
X_trans = selector.fit_transform(X=X, y=y, sample_weight=w)
print(f"The features going in the selector are : {selector.feature_names_in_}")
print(f"The support is : {selector.support_}")
print(f"The selected features are : {selector.get_feature_names_out()}")
The features going in the selector are : ['var0' 'var1' 'var2' 'var3' 'var4' 'var5' 'var6' 'var7' 'var8' 'var9'
'var10' 'var11' 'var12' 'nice_guys']
The support is : [ True True True True True True False False True False False True
True False]
The selected features are : ['var0' 'var1' 'var2' 'var3' 'var4' 'var5' 'var8' 'var11' 'var12']
[26]:
selector.plot_importance(log=False, style=None)
enable fasttreeshap implementation
[27]:
lgb_kwargs = {"objective": "rmse", "zero_as_missing": False}
selector = arfsfs.VariableImportance(
verbose=2, threshold=0.99, lgb_kwargs=lgb_kwargs, fastshap=True
)
X_trans = selector.fit_transform(X=X, y=y, sample_weight=w)
print(f"The features going in the selector are : {selector.feature_names_in_}")
print(f"The support is : {selector.support_}")
print(f"The selected features are : {selector.get_feature_names_out()}")
The features going in the selector are : ['var0' 'var1' 'var2' 'var3' 'var4' 'var5' 'var6' 'var7' 'var8' 'var9'
'var10' 'var11' 'var12' 'nice_guys']
The support is : [ True True True True True True False False True False False True
True False]
The selected features are : ['var0' 'var1' 'var2' 'var3' 'var4' 'var5' 'var8' 'var11' 'var12']
[28]:
selector.plot_importance(log=False, style=None)
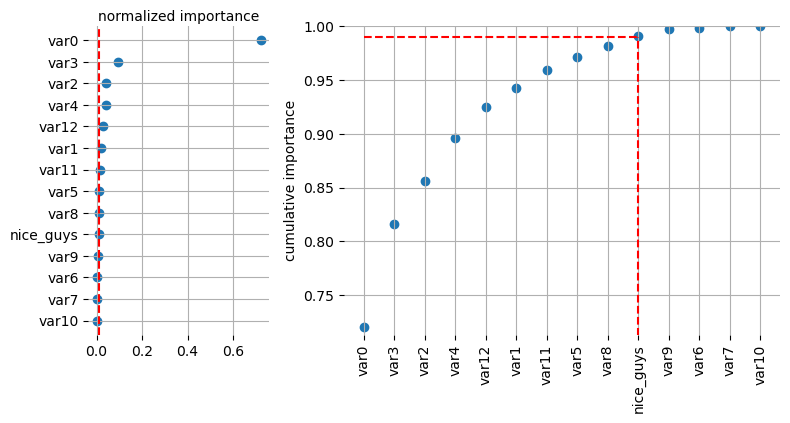
All at once - Sklearn Pipeline#
The selectors follow the scikit-learn base class, therefore they are compatible with scikit-learn in general. This has several advantages: - easier to maintain - easier to version - more flexible - running faster by removing unnecessary columns before going to the computational demanding steps
[29]:
from arfs.preprocessing import dtype_column_selector
cat_features_selector = dtype_column_selector(
dtype_include=["category", "object", "bool"],
dtype_exclude=[np.number],
pattern=None,
exclude_cols=["nice_guys"],
)
cat_features_selector(X)
[29]:
['var11']
[30]:
from sklearn.pipeline import Pipeline
from arfs.preprocessing import OrdinalEncoderPandas
encoder = Pipeline(
[
('zero_variance', arfsfs.UniqueValuesThreshold()),
("collinearity", arfsfs.CollinearityThreshold(threshold=0.75)),
("encoder", OrdinalEncoderPandas()),
]
)
X_encoded = encoder.fit(X).transform(X)
[31]:
encoder.fit(X)
[31]:
Pipeline(steps=[('zero_variance', UniqueValuesThreshold()),
('collinearity', CollinearityThreshold(threshold=0.75)),
('encoder', OrdinalEncoderPandas())])In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook. On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
Pipeline(steps=[('zero_variance', UniqueValuesThreshold()),
('collinearity', CollinearityThreshold(threshold=0.75)),
('encoder', OrdinalEncoderPandas())])UniqueValuesThreshold()
CollinearityThreshold(threshold=0.75)
OrdinalEncoderPandas()
[32]:
X_encoded = OrdinalEncoderPandas().fit_transform(X)
X_encoded
[32]:
| var0 | var1 | var2 | var3 | var4 | var5 | var6 | var7 | var8 | var9 | var10 | var11 | var12 | nice_guys | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1.016361 | 0.821664 | -1.184095 | 0.985738 | 1.050445 | -0.494190 | 2 | 0 | -0.361717 | 1 | 1.0 | 0.0 | 1.705980 | 4.0 |
| 1 | 0.929581 | 0.334013 | -1.063030 | 0.819273 | 1.016252 | 0.283845 | 1 | 0 | 0.178670 | 2 | 1.0 | 1.0 | NaN | 21.0 |
| 2 | 1.095456 | 0.187234 | -1.488666 | 1.087443 | 1.140542 | 0.962503 | 2 | 0 | -3.375579 | 2 | 1.0 | 2.0 | NaN | 19.0 |
| 3 | 1.318165 | 0.994528 | -1.370624 | 1.592398 | 1.315021 | 1.165595 | 0 | 0 | -0.449650 | 2 | 1.0 | 3.0 | 2.929707 | 25.0 |
| 4 | 0.849496 | 0.184859 | -0.806604 | 0.865702 | 0.991916 | -0.058833 | 1 | 0 | 0.763903 | 1 | 1.0 | 4.0 | NaN | 32.0 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 995 | 0.904073 | 0.009764 | -0.909486 | 0.831147 | 1.041658 | 0.081291 | 0 | 0 | -0.542799 | 3 | 1.0 | 495.0 | NaN | 10.0 |
| 996 | 1.370908 | 1.144173 | -1.129008 | 2.002148 | 1.418488 | 0.484591 | 0 | 1 | -1.932372 | 1 | 1.0 | 496.0 | 2.587139 | 0.0 |
| 997 | 1.113773 | 0.206798 | -1.371649 | 1.167287 | 1.079760 | 0.267475 | 0 | 0 | -0.329798 | 0 | 1.0 | 497.0 | 3.880225 | 17.0 |
| 998 | 0.905654 | 0.598419 | -0.792174 | 0.783557 | 1.275266 | 0.380826 | 1 | 0 | -1.987310 | 1 | 1.0 | 498.0 | 1.888725 | 17.0 |
| 999 | 1.138102 | 0.143818 | -1.348573 | 1.321900 | 1.309521 | 1.342722 | 1 | 0 | -0.871590 | 1 | 1.0 | 499.0 | NaN | 9.0 |
1000 rows × 14 columns
[33]:
basic_fs_pipeline = Pipeline(
[
("missing", arfsfs.MissingValueThreshold(threshold=0.05)),
("unique", arfsfs.UniqueValuesThreshold(threshold=1)),
("cardinality", arfsfs.CardinalityThreshold(threshold=10)),
("collinearity", arfsfs.CollinearityThreshold(threshold=0.75)),
("encoder", OrdinalEncoderPandas()),
(
"lowimp",
arfsfs.VariableImportance(
verbose=2, threshold=0.99, lgb_kwargs=lgb_kwargs, encode=False
),
),
]
)
X_trans = basic_fs_pipeline.fit_transform(
X=X, y=y, collinearity__sample_weight=w, lowimp__sample_weight=w
)
[34]:
basic_fs_pipeline
[34]:
Pipeline(steps=[('missing', MissingValueThreshold()),
('unique', UniqueValuesThreshold()),
('cardinality', CardinalityThreshold(threshold=10)),
('collinearity', CollinearityThreshold(threshold=0.75)),
('encoder', OrdinalEncoderPandas()),
('lowimp',
VariableImportance(encode=False,
lgb_kwargs={'metric': 'rmse',
'objective': 'rmse',
'verbosity': -1,
'zero_as_missing': False},
verbose=2))])In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook. On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
Pipeline(steps=[('missing', MissingValueThreshold()),
('unique', UniqueValuesThreshold()),
('cardinality', CardinalityThreshold(threshold=10)),
('collinearity', CollinearityThreshold(threshold=0.75)),
('encoder', OrdinalEncoderPandas()),
('lowimp',
VariableImportance(encode=False,
lgb_kwargs={'metric': 'rmse',
'objective': 'rmse',
'verbosity': -1,
'zero_as_missing': False},
verbose=2))])MissingValueThreshold()
UniqueValuesThreshold()
CardinalityThreshold(threshold=10)
CollinearityThreshold(threshold=0.75)
OrdinalEncoderPandas()
VariableImportance(encode=False,
lgb_kwargs={'metric': 'rmse', 'objective': 'rmse',
'verbosity': -1, 'zero_as_missing': False},
verbose=2)[35]:
# X_trans = basic_fs_pipeline.transform(X)
X_trans.head()
[35]:
| var1 | var2 | var3 | var4 | var5 | var8 | |
|---|---|---|---|---|---|---|
| 0 | 0.821664 | -1.184095 | 0.985738 | 1.050445 | -0.494190 | -0.361717 |
| 1 | 0.334013 | -1.063030 | 0.819273 | 1.016252 | 0.283845 | 0.178670 |
| 2 | 0.187234 | -1.488666 | 1.087443 | 1.140542 | 0.962503 | -3.375579 |
| 3 | 0.994528 | -1.370624 | 1.592398 | 1.315021 | 1.165595 | -0.449650 |
| 4 | 0.184859 | -0.806604 | 0.865702 | 0.991916 | -0.058833 | 0.763903 |
[36]:
type(basic_fs_pipeline.named_steps["encoder"])
[36]:
arfs.preprocessing.OrdinalEncoderPandas
[37]:
arfsfs.make_fs_summary(basic_fs_pipeline)
[37]:
| predictor | missing | unique | cardinality | collinearity | encoder | lowimp | |
|---|---|---|---|---|---|---|---|
| 0 | var0 | 1 | 1 | 1 | 0 | nan | nan |
| 1 | var1 | 1 | 1 | 1 | 1 | nan | 1 |
| 2 | var2 | 1 | 1 | 1 | 1 | nan | 1 |
| 3 | var3 | 1 | 1 | 1 | 1 | nan | 1 |
| 4 | var4 | 1 | 1 | 1 | 1 | nan | 1 |
| 5 | var5 | 1 | 1 | 1 | 1 | nan | 1 |
| 6 | var6 | 1 | 1 | 1 | 1 | nan | 0 |
| 7 | var7 | 1 | 1 | 1 | 1 | nan | 0 |
| 8 | var8 | 1 | 1 | 1 | 1 | nan | 1 |
| 9 | var9 | 1 | 1 | 1 | 1 | nan | 0 |
| 10 | var10 | 1 | 0 | nan | nan | nan | nan |
| 11 | var11 | 1 | 1 | 0 | nan | nan | nan |
| 12 | var12 | 0 | nan | nan | nan | nan | nan |
| 13 | nice_guys | 1 | 1 | 0 | nan | nan | nan |
Using the cancer data
[38]:
import arfs
print(f"ARFS version {arfs.__version__}")
from arfs.utils import load_data
cancer = load_data(name="cancer")
X, y = cancer.data, cancer.target
y = y.astype(int)
selector = arfsfs.CollinearityThreshold(threshold=0.85, n_jobs=1)
X_trans = selector.fit_transform(X)
print(f"The features going in the selector are : {selector.feature_names_in_}")
print(f"The support is : {selector.support_}")
print(f"The selected features are : {selector.get_feature_names_out()}")
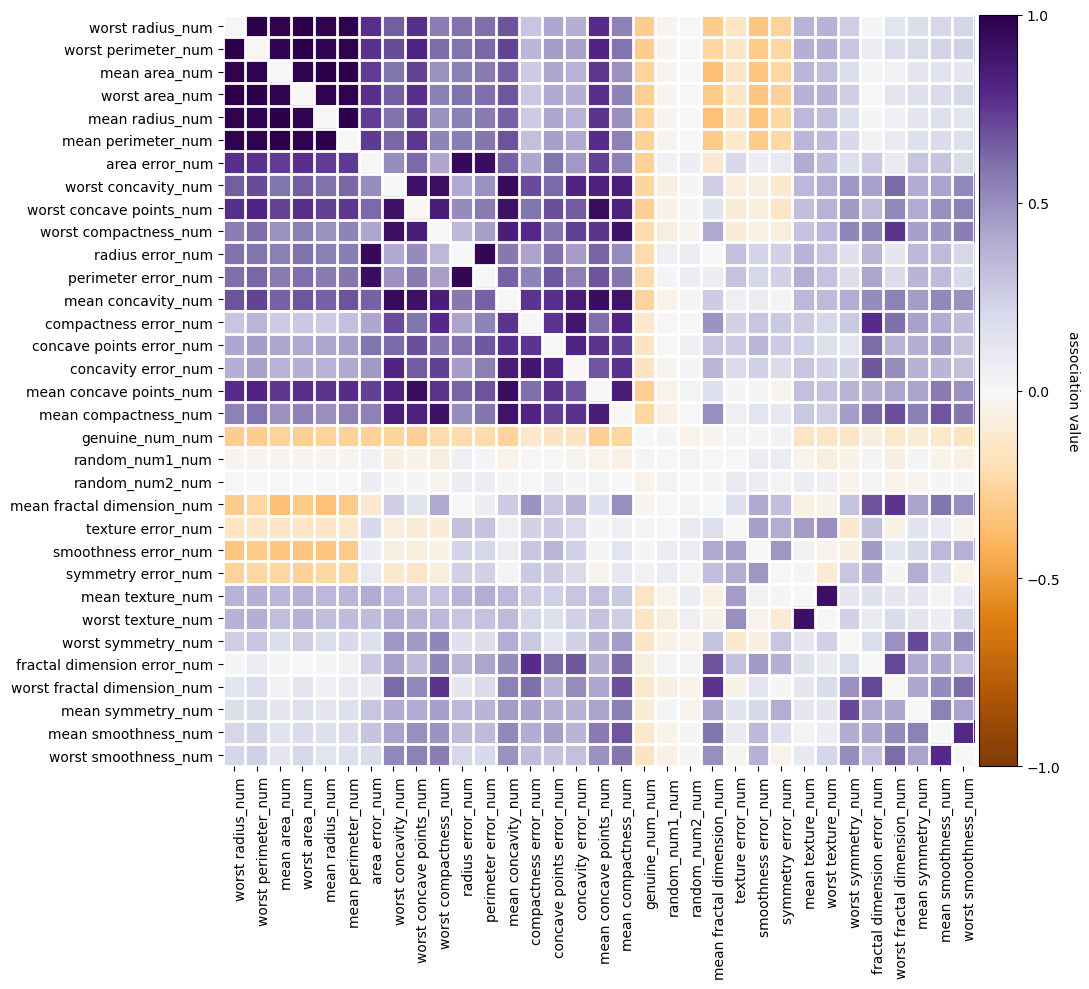
f = selector.plot_association()
ARFS version 2.2.3
The features going in the selector are : ['mean radius' 'mean texture' 'mean perimeter' 'mean area'
'mean smoothness' 'mean compactness' 'mean concavity'
'mean concave points' 'mean symmetry' 'mean fractal dimension'
'radius error' 'texture error' 'perimeter error' 'area error'
'smoothness error' 'compactness error' 'concavity error'
'concave points error' 'symmetry error' 'fractal dimension error'
'worst radius' 'worst texture' 'worst perimeter' 'worst area'
'worst smoothness' 'worst compactness' 'worst concavity'
'worst concave points' 'worst symmetry' 'worst fractal dimension'
'random_num1' 'random_num2' 'genuine_num']
The support is : [False True False True True False False True True True True True
False False True False True True True True False False False False
True True False False True True True True True]
The selected features are : ['mean texture' 'mean area' 'mean smoothness' 'mean concave points'
'mean symmetry' 'mean fractal dimension' 'radius error' 'texture error'
'smoothness error' 'concavity error' 'concave points error'
'symmetry error' 'fractal dimension error' 'worst smoothness'
'worst compactness' 'worst symmetry' 'worst fractal dimension'
'random_num1' 'random_num2' 'genuine_num']
[39]:
selector.not_selected_features_
[39]:
array(['mean radius', 'mean perimeter', 'mean compactness',
'mean concavity', 'perimeter error', 'area error',
'compactness error', 'worst radius', 'worst texture',
'worst perimeter', 'worst area', 'worst concavity',
'worst concave points'], dtype=object)
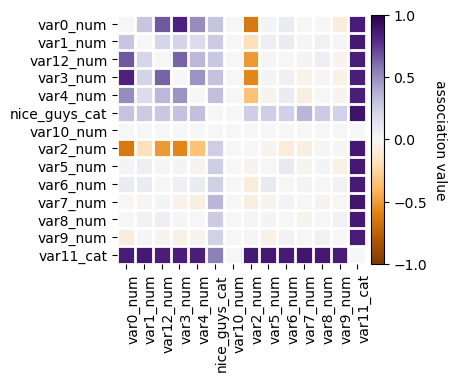
Association of the non-selected features with all the input features
[40]:
def highlight_gt_threshold(v, props='', threshold=0.85):
return props if v > threshold else None
selector.assoc_matrix_.loc[
:, selector.not_selected_features_
].style.applymap(highlight_gt_threshold, props='color:red;', threshold=0.85)
[40]:
| mean radius | mean perimeter | mean compactness | mean concavity | perimeter error | area error | compactness error | worst radius | worst texture | worst perimeter | worst area | worst concavity | worst concave points | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| area error | 0.738077 | 0.745824 | 0.539511 | 0.644344 | 0.926937 | 0.000000 | 0.409830 | 0.774244 | 0.327857 | 0.768336 | 0.775662 | 0.500307 | 0.619539 |
| compactness error | 0.264904 | 0.308620 | 0.817875 | 0.761230 | 0.532081 | 0.409830 | 0.000000 | 0.285413 | 0.209979 | 0.344865 | 0.278844 | 0.701251 | 0.587471 |
| concave points error | 0.410576 | 0.441996 | 0.732425 | 0.774656 | 0.669574 | 0.588749 | 0.764150 | 0.410221 | 0.157304 | 0.448363 | 0.403587 | 0.624555 | 0.692071 |
| concavity error | 0.364555 | 0.402277 | 0.772283 | 0.858306 | 0.547805 | 0.461359 | 0.880965 | 0.381860 | 0.235945 | 0.432895 | 0.377836 | 0.811327 | 0.656814 |
| fractal dimension error | -0.008411 | 0.032429 | 0.621121 | 0.513593 | 0.420114 | 0.259401 | 0.781396 | 0.013324 | 0.083174 | 0.063012 | 0.007312 | 0.431677 | 0.331206 |
| genuine_num | -0.257916 | -0.266154 | -0.244416 | -0.265017 | -0.220325 | -0.268824 | -0.137252 | -0.289229 | -0.151634 | -0.290689 | -0.287949 | -0.253526 | -0.285553 |
| mean area | 0.999602 | 0.997068 | 0.488988 | 0.642557 | 0.568237 | 0.741518 | 0.260362 | 0.979258 | 0.318178 | 0.971822 | 0.980264 | 0.593736 | 0.723390 |
| mean compactness | 0.497578 | 0.543925 | 0.000000 | 0.896518 | 0.583520 | 0.539511 | 0.817875 | 0.542626 | 0.255305 | 0.592254 | 0.531590 | 0.837921 | 0.825473 |
| mean concave points | 0.759702 | 0.788629 | 0.848295 | 0.927352 | 0.679841 | 0.726982 | 0.608388 | 0.787411 | 0.300562 | 0.813960 | 0.780395 | 0.827281 | 0.937075 |
| mean concavity | 0.645728 | 0.681958 | 0.896518 | 0.000000 | 0.646199 | 0.644344 | 0.761230 | 0.682316 | 0.335866 | 0.722424 | 0.676628 | 0.938543 | 0.904938 |
| mean fractal dimension | -0.349931 | -0.304891 | 0.499195 | 0.258174 | 0.055309 | -0.120333 | 0.481139 | -0.294540 | -0.047791 | -0.247456 | -0.304927 | 0.242611 | 0.139152 |
| mean perimeter | 0.997802 | 0.000000 | 0.543925 | 0.681958 | 0.582789 | 0.745824 | 0.308620 | 0.981244 | 0.323109 | 0.978980 | 0.980864 | 0.632106 | 0.757526 |
| mean radius | 0.000000 | 0.997802 | 0.497578 | 0.645728 | 0.565520 | 0.738077 | 0.264904 | 0.978604 | 0.314911 | 0.971555 | 0.978863 | 0.596043 | 0.727265 |
| mean smoothness | 0.148510 | 0.182923 | 0.678806 | 0.518511 | 0.331360 | 0.296059 | 0.392455 | 0.203453 | 0.060645 | 0.226345 | 0.191735 | 0.429107 | 0.498868 |
| mean symmetry | 0.120242 | 0.150049 | 0.552203 | 0.446793 | 0.354888 | 0.288322 | 0.435714 | 0.164552 | 0.118890 | 0.190526 | 0.154462 | 0.394481 | 0.397477 |
| mean texture | 0.340956 | 0.348142 | 0.266499 | 0.342646 | 0.386813 | 0.395139 | 0.263591 | 0.366547 | 0.909218 | 0.375273 | 0.368335 | 0.339725 | 0.319235 |
| perimeter error | 0.565520 | 0.582789 | 0.583520 | 0.646199 | 0.000000 | 0.926937 | 0.532081 | 0.606902 | 0.302553 | 0.626896 | 0.605163 | 0.490340 | 0.569428 |
| radius error | 0.550247 | 0.560326 | 0.506582 | 0.575277 | 0.957728 | 0.952867 | 0.428186 | 0.598030 | 0.283581 | 0.592509 | 0.595732 | 0.404431 | 0.508662 |
| random_num1 | -0.022225 | -0.025336 | -0.057914 | -0.033234 | 0.022807 | 0.031618 | -0.004894 | -0.024181 | -0.073682 | -0.030910 | -0.024583 | -0.067753 | -0.050934 |
| random_num2 | 0.000498 | 0.002551 | 0.000041 | 0.008068 | 0.068938 | 0.055542 | 0.005724 | 0.003751 | 0.046919 | 0.006469 | 0.005232 | -0.007958 | 0.011413 |
| smoothness error | -0.326385 | -0.311147 | 0.127381 | 0.070321 | 0.209335 | 0.066969 | 0.285264 | -0.321112 | -0.036290 | -0.308749 | -0.323724 | -0.063848 | -0.076503 |
| symmetry error | -0.241376 | -0.228187 | 0.098388 | 0.022753 | 0.238855 | 0.093502 | 0.272750 | -0.261267 | -0.104702 | -0.246712 | -0.266705 | -0.118144 | -0.140795 |
| texture error | -0.144499 | -0.137578 | 0.047766 | 0.051318 | 0.302979 | 0.200930 | 0.230048 | -0.148594 | 0.496551 | -0.142855 | -0.147786 | -0.070625 | -0.097025 |
| worst area | 0.978863 | 0.980864 | 0.531590 | 0.676628 | 0.605163 | 0.775662 | 0.278844 | 0.998891 | 0.372376 | 0.992433 | 0.000000 | 0.651120 | 0.773945 |
| worst compactness | 0.491357 | 0.534565 | 0.901029 | 0.849985 | 0.438416 | 0.413658 | 0.789431 | 0.558316 | 0.342319 | 0.613070 | 0.550007 | 0.914894 | 0.844454 |
| worst concave points | 0.727265 | 0.757526 | 0.825473 | 0.904938 | 0.569428 | 0.619539 | 0.587471 | 0.780632 | 0.365309 | 0.812983 | 0.773945 | 0.902301 | 0.000000 |
| worst concavity | 0.596043 | 0.632106 | 0.837921 | 0.938543 | 0.490340 | 0.500307 | 0.701251 | 0.655942 | 0.387009 | 0.700572 | 0.651120 | 0.000000 | 0.902301 |
| worst fractal dimension | 0.044564 | 0.088961 | 0.688986 | 0.541838 | 0.185534 | 0.091670 | 0.604844 | 0.127449 | 0.193191 | 0.179003 | 0.118734 | 0.623128 | 0.516664 |
| worst perimeter | 0.971555 | 0.978980 | 0.592254 | 0.722424 | 0.626896 | 0.768336 | 0.344865 | 0.993548 | 0.381022 | 0.000000 | 0.992433 | 0.700572 | 0.812983 |
| worst radius | 0.978604 | 0.981244 | 0.542626 | 0.682316 | 0.606902 | 0.774244 | 0.285413 | 0.000000 | 0.371230 | 0.993548 | 0.998891 | 0.655942 | 0.780632 |
| worst smoothness | 0.125789 | 0.156611 | 0.578902 | 0.488775 | 0.197899 | 0.188777 | 0.320500 | 0.218616 | 0.217799 | 0.241172 | 0.210063 | 0.519490 | 0.543982 |
| worst symmetry | 0.174698 | 0.199007 | 0.450333 | 0.383667 | 0.166606 | 0.154415 | 0.265987 | 0.257165 | 0.226816 | 0.281383 | 0.248358 | 0.476179 | 0.460711 |
| worst texture | 0.314911 | 0.323109 | 0.255305 | 0.335866 | 0.302553 | 0.327857 | 0.209979 | 0.371230 | 0.000000 | 0.381022 | 0.372376 | 0.387009 | 0.365309 |
Association of the selected features only
[41]:
selector.assoc_matrix_.loc[
selector.selected_features_, selector.selected_features_
].style.applymap(highlight_gt_threshold, props='color:red;', threshold=0.85)
[41]:
| mean texture | mean area | mean smoothness | mean concave points | mean symmetry | mean fractal dimension | radius error | texture error | smoothness error | concavity error | concave points error | symmetry error | fractal dimension error | worst smoothness | worst compactness | worst symmetry | worst fractal dimension | random_num1 | random_num2 | genuine_num | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| mean texture | 0.000000 | 0.344145 | 0.024649 | 0.306891 | 0.110130 | -0.059303 | 0.363621 | 0.450720 | 0.037048 | 0.287188 | 0.238610 | 0.008945 | 0.147605 | 0.101401 | 0.290917 | 0.120693 | 0.116144 | -0.033471 | 0.063706 | -0.148910 |
| mean area | 0.344145 | 0.000000 | 0.138053 | 0.755165 | 0.113928 | -0.358425 | 0.553388 | -0.142469 | -0.327431 | 0.362308 | 0.406468 | -0.243507 | -0.012688 | 0.119712 | 0.485813 | 0.170860 | 0.038758 | -0.020784 | 0.000305 | -0.258352 |
| mean smoothness | 0.024649 | 0.138053 | 0.000000 | 0.565172 | 0.542228 | 0.588465 | 0.334282 | 0.091283 | 0.338692 | 0.354730 | 0.438826 | 0.150740 | 0.413429 | 0.796085 | 0.481384 | 0.393579 | 0.511457 | -0.043211 | 0.008687 | -0.137660 |
| mean concave points | 0.306891 | 0.755165 | 0.565172 | 0.000000 | 0.423767 | 0.142659 | 0.635054 | 0.008710 | 0.016798 | 0.674668 | 0.758438 | -0.028353 | 0.378374 | 0.490035 | 0.758309 | 0.355477 | 0.421110 | -0.039682 | 0.025712 | -0.288981 |
| mean symmetry | 0.110130 | 0.113928 | 0.542228 | 0.423767 | 0.000000 | 0.428467 | 0.337912 | 0.139124 | 0.206106 | 0.367637 | 0.382736 | 0.384123 | 0.402630 | 0.424230 | 0.440828 | 0.710359 | 0.410069 | 0.017243 | -0.016538 | -0.096999 |
| mean fractal dimension | -0.059303 | -0.358425 | 0.588465 | 0.142659 | 0.428467 | 0.000000 | 0.001477 | 0.157103 | 0.401530 | 0.344007 | 0.286393 | 0.314165 | 0.683800 | 0.493474 | 0.403653 | 0.295046 | 0.760771 | 0.001102 | -0.012412 | -0.019863 |
| radius error | 0.363621 | 0.553388 | 0.334282 | 0.635054 | 0.337912 | 0.001477 | 0.000000 | 0.309672 | 0.223469 | 0.452542 | 0.595594 | 0.240118 | 0.348164 | 0.203760 | 0.339725 | 0.147213 | 0.111043 | 0.054287 | 0.059528 | -0.224582 |
| texture error | 0.450720 | -0.142469 | 0.091283 | 0.008710 | 0.139124 | 0.157103 | 0.309672 | 0.000000 | 0.443640 | 0.189277 | 0.261263 | 0.389080 | 0.309209 | -0.023095 | -0.090069 | -0.119890 | -0.048143 | 0.000659 | 0.082678 | 0.021685 |
| smoothness error | 0.037048 | -0.327431 | 0.338692 | 0.016798 | 0.206106 | 0.401530 | 0.223469 | 0.443640 | 0.000000 | 0.236961 | 0.345073 | 0.473579 | 0.460478 | 0.372247 | -0.049245 | -0.067149 | 0.129752 | 0.076456 | 0.075758 | 0.010950 |
| concavity error | 0.287188 | 0.362308 | 0.354730 | 0.674668 | 0.367637 | 0.344007 | 0.452542 | 0.189277 | 0.236961 | 0.000000 | 0.804773 | 0.191862 | 0.668224 | 0.305368 | 0.731517 | 0.230690 | 0.505962 | -0.024256 | 0.014859 | -0.161208 |
| concave points error | 0.238610 | 0.406468 | 0.438826 | 0.758438 | 0.382736 | 0.286393 | 0.595594 | 0.261263 | 0.345073 | 0.804773 | 0.000000 | 0.262155 | 0.611329 | 0.294840 | 0.585929 | 0.132782 | 0.357090 | 0.006869 | 0.052780 | -0.162009 |
| symmetry error | 0.008945 | -0.243507 | 0.150740 | -0.028353 | 0.384123 | 0.314165 | 0.240118 | 0.389080 | 0.473579 | 0.191862 | 0.262155 | 0.000000 | 0.380711 | -0.042873 | -0.082054 | 0.283201 | 0.011133 | 0.073219 | 0.016392 | 0.035264 |
| fractal dimension error | 0.147605 | -0.012688 | 0.413429 | 0.378374 | 0.402630 | 0.683800 | 0.348164 | 0.309209 | 0.460478 | 0.668224 | 0.611329 | 0.380711 | 0.000000 | 0.312293 | 0.526899 | 0.172926 | 0.712771 | 0.015924 | 0.019052 | -0.073278 |
| worst smoothness | 0.101401 | 0.119712 | 0.796085 | 0.490035 | 0.424230 | 0.493474 | 0.203760 | -0.023095 | 0.372247 | 0.305368 | 0.294840 | -0.042873 | 0.312293 | 0.000000 | 0.560156 | 0.501230 | 0.614796 | -0.060299 | 0.011472 | -0.158576 |
| worst compactness | 0.290917 | 0.485813 | 0.481384 | 0.758309 | 0.440828 | 0.403653 | 0.339725 | -0.090069 | -0.049245 | 0.731517 | 0.585929 | -0.082054 | 0.526899 | 0.560156 | 0.000000 | 0.527102 | 0.762247 | -0.071538 | -0.023894 | -0.224983 |
| worst symmetry | 0.120693 | 0.170860 | 0.393579 | 0.355477 | 0.710359 | 0.295046 | 0.147213 | -0.119890 | -0.067149 | 0.230690 | 0.132782 | 0.283201 | 0.172926 | 0.501230 | 0.527102 | 0.000000 | 0.488439 | -0.048632 | -0.032499 | -0.143367 |
| worst fractal dimension | 0.116144 | 0.038758 | 0.511457 | 0.421110 | 0.410069 | 0.760771 | 0.111043 | -0.048143 | 0.129752 | 0.505962 | 0.357090 | 0.011133 | 0.712771 | 0.614796 | 0.762247 | 0.488439 | 0.000000 | -0.063255 | -0.037140 | -0.137431 |
| random_num1 | -0.033471 | -0.020784 | -0.043211 | -0.039682 | 0.017243 | 0.001102 | 0.054287 | 0.000659 | 0.076456 | -0.024256 | 0.006869 | 0.073219 | 0.015924 | -0.060299 | -0.071538 | -0.048632 | -0.063255 | 0.000000 | 0.025853 | 0.016705 |
| random_num2 | 0.063706 | 0.000305 | 0.008687 | 0.025712 | -0.016538 | -0.012412 | 0.059528 | 0.082678 | 0.075758 | 0.014859 | 0.052780 | 0.016392 | 0.019052 | 0.011472 | -0.023894 | -0.032499 | -0.037140 | 0.025853 | 0.000000 | -0.043352 |
| genuine_num | -0.148910 | -0.258352 | -0.137660 | -0.288981 | -0.096999 | -0.019863 | -0.224582 | 0.021685 | 0.010950 | -0.161208 | -0.162009 | 0.035264 | -0.073278 | -0.158576 | -0.224983 | -0.143367 | -0.137431 | 0.016705 | -0.043352 | 0.000000 |
[50]:
%timeit -n1 pass
res = arfs.association.wcorr_matrix(X, sample_weight=None, n_jobs=-1, handle_na='drop', method='pearson')
251 ns ± 242 ns per loop (mean ± std. dev. of 7 runs, 1 loop each)
[48]:
%timeit -n1 pass
from scipy import stats
res = stats.spearmanr(X)
137 ns ± 89 ns per loop (mean ± std. dev. of 7 runs, 1 loop each)
[ ]: