ARFS with Categorical predictors#
Let’s illustrate how ARFS is handling the categorical predictors
[1]:
# from IPython.core.display import display, HTML
# display(HTML("<style>.container { width:95% !important; }</style>"))
import catboost
import numpy as np
import pandas as pd
import matplotlib as mpl
import matplotlib.pyplot as plt
import gc
import shap
from boruta import BorutaPy as bp
from sklearn.datasets import fetch_openml
from sklearn.inspection import permutation_importance
from sklearn.pipeline import Pipeline
from sklearn.datasets import fetch_openml
from sklearn.inspection import permutation_importance
from sklearn.base import clone
from sklearn.ensemble import RandomForestRegressor, RandomForestClassifier
from lightgbm import LGBMRegressor, LGBMClassifier
from xgboost import XGBRegressor, XGBClassifier
from catboost import CatBoostRegressor, CatBoostClassifier
from sys import getsizeof, path
import arfs
import arfs.feature_selection as arfsfs
import arfs.feature_selection.allrelevant as arfsgroot
from arfs.utils import LightForestClassifier, LightForestRegressor
from arfs.benchmark import highlight_tick, compare_varimp, sklearn_pimp_bench
from arfs.utils import load_data
# plt.style.use('fivethirtyeight')
rng = np.random.RandomState(seed=42)
# import warnings
# warnings.filterwarnings('ignore')
Using `tqdm.autonotebook.tqdm` in notebook mode. Use `tqdm.tqdm` instead to force console mode (e.g. in jupyter console)
[2]:
print(f"Run with ARFS {arfs.__version__}")
Run with ARFS 2.0.5
[3]:
%matplotlib inline
[4]:
gc.enable()
gc.collect()
[4]:
4
Simple Usage#
Most of the tree-based models do not handle categorical predictors without pre-processing. In the three different All Relevant Feature Selection methods, the categorical predictors are handled automatically. How?
the non-numeric columns are selected
integer encoding is performed
for lightGBM and XGBoost, the columns are encoded using contiguous integers and are passed as categoricals. See the official documentation
for CatBoost, the columns are encoded but the categorical columns are passed to the fit methods
estimator.fit(X_tr, y_tr, sample_weight=w_tr, cat_features=obj_feat)(for lightGBM, this param is pass to the estimator not the fit method).
This is done internally and automatically, you don’t have to set it yourself (if you do so, it’ll throw an error).
Let’s see an example using the Titanic data, with synthetic predictors added for benchmarking purpose.
[5]:
titanic = load_data(name="Titanic")
X, y = titanic.data, titanic.target
y = y.astype(int)
The default value of `parser` will change from `'liac-arff'` to `'auto'` in 1.4. You can set `parser='auto'` to silence this warning. Therefore, an `ImportError` will be raised from 1.4 if the dataset is dense and pandas is not installed. Note that the pandas parser may return different data types. See the Notes Section in fetch_openml's API doc for details.
[6]:
X.dtypes
[6]:
pclass object
sex object
embarked object
random_cat object
is_alone object
title object
age float64
family_size float64
fare float64
random_num float64
dtype: object
[7]:
X.head()
[7]:
| pclass | sex | embarked | random_cat | is_alone | title | age | family_size | fare | random_num | |
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1.0 | female | S | Fry | 1 | Mrs | 29.0000 | 0.0 | 211.3375 | 0.496714 |
| 1 | 1.0 | male | S | Bender | 0 | Master | 0.9167 | 3.0 | 151.5500 | -0.138264 |
| 2 | 1.0 | female | S | Thanos | 0 | Mrs | 2.0000 | 3.0 | 151.5500 | 0.647689 |
| 3 | 1.0 | male | S | Morty | 0 | Mr | 30.0000 | 3.0 | 151.5500 | 1.523030 |
| 4 | 1.0 | female | S | Morty | 0 | Mrs | 25.0000 | 3.0 | 151.5500 | -0.234153 |
GrootCV#
[8]:
%%time
# GrootCV
feat_selector = arfsgroot.GrootCV(
objective="binary", cutoff=1, n_folds=5, n_iter=5, silent=True
)
feat_selector.fit(X, y, sample_weight=None)
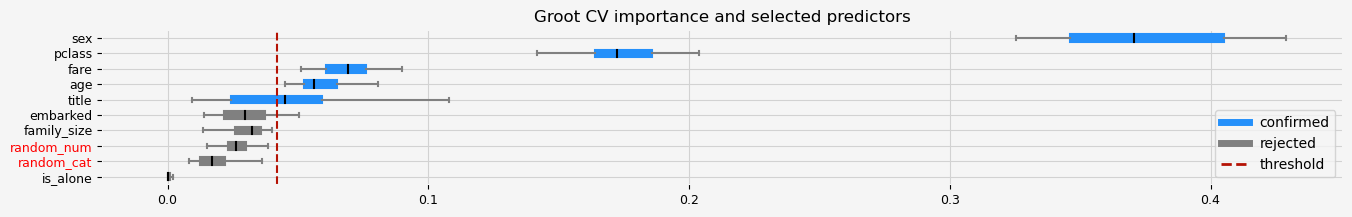
print(f"The selected features: {feat_selector.get_feature_names_out()}")
print(f"The agnostic ranking: {feat_selector.ranking_}")
print(f"The naive ranking: {feat_selector.ranking_absolutes_}")
fig = feat_selector.plot_importance(n_feat_per_inch=5)
# highlight synthetic random variable
fig = highlight_tick(figure=fig, str_match="random")
fig = highlight_tick(figure=fig, str_match="genuine", color="green")
plt.show()
The selected features: ['pclass' 'sex' 'title' 'age' 'fare']
The agnostic ranking: [2 2 1 1 1 2 2 1 2 1]
The naive ranking: ['sex', 'pclass', 'fare', 'age', 'title', 'embarked', 'family_size', 'random_num', 'random_cat', 'is_alone']
CPU times: user 18.5 s, sys: 375 ms, total: 18.8 s
Wall time: 5.76 s
BoostAGroota#
[9]:
# be sure to use the same but non-fitted estimator
model = LGBMClassifier(random_state=1258, verbose=-1, n_estimators=100)
# BoostAGroota
feat_selector = arfsgroot.BoostAGroota(
estimator=model, cutoff=1, iters=10, max_rounds=10, delta=0.1, importance="fastshap"
)
feat_selector.fit(X, y, sample_weight=None)
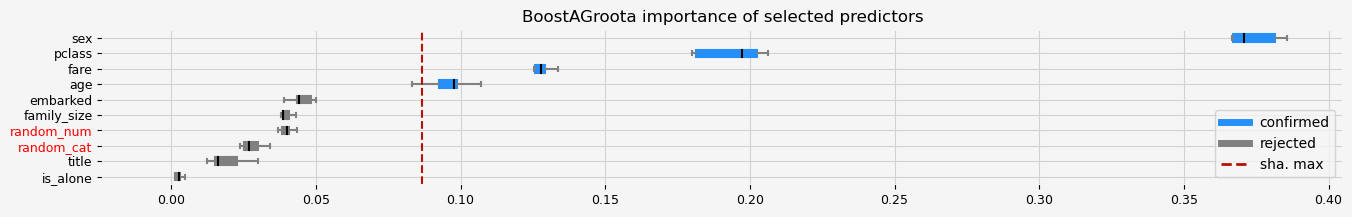
print(f"The selected features: {feat_selector.get_feature_names_out()}")
print(f"The agnostic ranking: {feat_selector.ranking_}")
print(f"The naive ranking: {feat_selector.ranking_absolutes_}")
fig = feat_selector.plot_importance(n_feat_per_inch=5)
# highlight synthetic random variable
fig = highlight_tick(figure=fig, str_match="random")
fig = highlight_tick(figure=fig, str_match="genuine", color="green")
plt.show()
The selected features: ['pclass' 'sex' 'age' 'fare']
The agnostic ranking: [2 2 1 1 1 1 2 1 2 1]
The naive ranking: ['sex', 'pclass', 'fare', 'age', 'embarked', 'family_size', 'random_num', 'random_cat', 'title', 'is_alone']
Leshy#
You don’t have to worry about the encoding, everything is performed internally. Even if you used CatBoost, the categorical predictors are handled internally as well.
[10]:
cat_idx = [X.columns.get_loc(col) for col in titanic.categorical]
[11]:
%%time
# Leshy
model = LGBMClassifier(random_state=42, verbose=-1, n_estimators=100)
# Leshy, all the predictors, no-preprocessing
feat_selector = arfsgroot.Leshy(
model,
n_estimators=1000,
verbose=1,
max_iter=10,
random_state=42,
importance="fastshap",
)
feat_selector.fit(X, y, sample_weight=None)
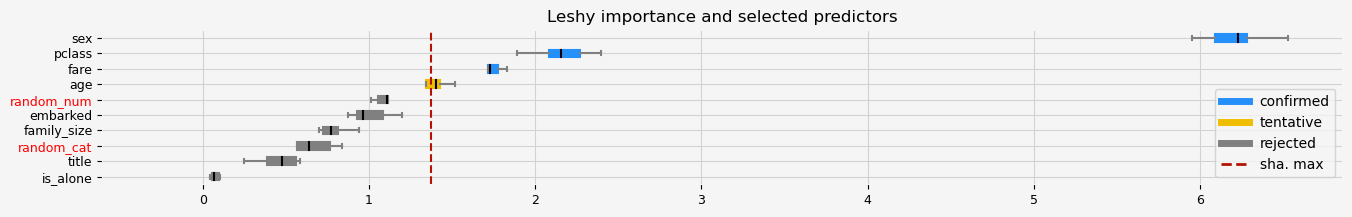
print(f"The selected features: {feat_selector.get_feature_names_out()}")
print(f"The agnostic ranking: {feat_selector.ranking_}")
print(f"The naive ranking: {feat_selector.ranking_absolutes_}")
fig = feat_selector.plot_importance(n_feat_per_inch=5)
# highlight synthetic random variable
fig = highlight_tick(figure=fig, str_match="random")
fig = highlight_tick(figure=fig, str_match="genuine", color="green")
plt.show()
Leshy finished running using native var. imp.
Iteration: 1 / 10
Confirmed: 3
Tentative: 1
Rejected: 6
All relevant predictors selected in 00:00:18.75
The selected features: ['pclass' 'sex' 'fare']
The agnostic ranking: [1 1 4 6 8 7 2 5 1 3]
The naive ranking: ['sex', 'pclass', 'fare', 'age', 'random_num', 'embarked', 'family_size', 'random_cat', 'title', 'is_alone']
CPU times: user 38.4 s, sys: 1.04 s, total: 39.5 s
Wall time: 19.3 s
For CatBoost, you just have to configure the estimator without taking care of the categorical predictors. They will be passed to the fit method internally, as illustrated in the code snippet of the ARFS package
[...]
obj_feat = list(set(list(X_tr.columns)) - set(list(X_tr.select_dtypes(include=[np.number]))))
[...]
if check_if_tree_based(estimator):
try:
# handle cat features if supported by the fit method
if is_catboost(estimator) or ('cat_feature' in estimator.fit.__code__.co_varnames):
model = estimator.fit(X_tr, y_tr, sample_weight=w_tr, cat_features=cat_idx)
else:
model = estimator.fit(X_tr, y_tr, sample_weight=w_tr)
[...]
[12]:
%%time
model = CatBoostClassifier(random_state=42, verbose=0)
# Leshy, all the predictors, no-preprocessing
feat_selector = arfsgroot.Leshy(
model,
n_estimators=100,
verbose=1,
max_iter=10,
random_state=42,
importance="fastshap",
)
feat_selector.fit(X, y, sample_weight=None)
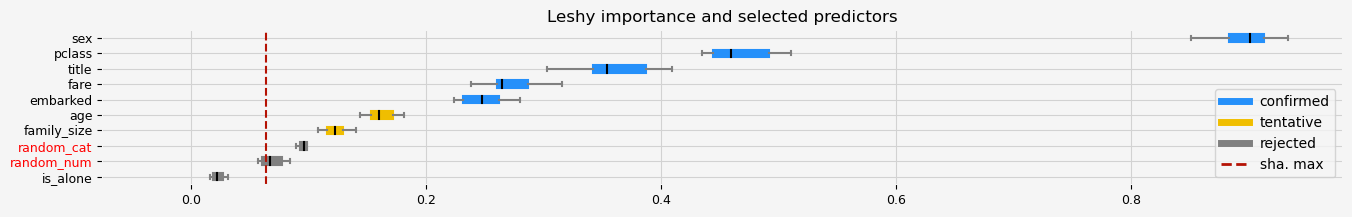
print(f"The selected features: {feat_selector.get_feature_names_out()}")
print(f"The agnostic ranking: {feat_selector.ranking_}")
print(f"The naive ranking: {feat_selector.ranking_absolutes_}")
fig = feat_selector.plot_importance(n_feat_per_inch=5)
# highlight synthetic random variable
fig = highlight_tick(figure=fig, str_match="random")
fig = highlight_tick(figure=fig, str_match="genuine", color="green")
plt.show()
Leshy finished running using native var. imp.
Iteration: 1 / 10
Confirmed: 5
Tentative: 2
Rejected: 3
All relevant predictors selected in 00:00:03.45
The selected features: ['pclass' 'sex' 'embarked' 'title' 'fare']
The agnostic ranking: [1 1 1 3 5 1 2 2 1 4]
The naive ranking: ['sex', 'pclass', 'title', 'fare', 'embarked', 'age', 'family_size', 'random_cat', 'random_num', 'is_alone']
CPU times: user 8.09 s, sys: 1.02 s, total: 9.11 s
Wall time: 3.83 s