ARFS - Customizing GrootCV#
GrootCV allows users to customize certain parameters when using the lightGBM algorithm in Python. However, it’s essential to be aware that some of these parameters will be automatically adjusted by the algorithm itself. Specifically, the overridden parameters are objective, verbose, is_balanced, and n_iterations.
[1]:
# from IPython.core.display import display, HTML
# display(HTML("<style>.container { width:95% !important; }</style>"))
import time
import numpy as np
import pandas as pd
import matplotlib as mpl
import matplotlib.pyplot as plt
import arfs
from arfs.feature_selection import GrootCV
from arfs.utils import load_data
from arfs.benchmark import highlight_tick
rng = np.random.RandomState(seed=42)
# import warnings
# warnings.filterwarnings('ignore')
Using `tqdm.autonotebook.tqdm` in notebook mode. Use `tqdm.tqdm` instead to force console mode (e.g. in jupyter console)
[2]:
import os
import multiprocessing
def get_physical_cores():
if os.name == "posix": # For Unix-based systems (e.g., Linux, macOS)
try:
return os.sysconf("SC_NPROCESSORS_ONLN")
except ValueError:
pass
elif os.name == "nt": # For Windows
try:
return int(os.environ["NUMBER_OF_PROCESSORS"])
except (ValueError, KeyError):
pass
return multiprocessing.cpu_count()
num_physical_cores = get_physical_cores()
print(f"Number of physical cores: {num_physical_cores}")
Number of physical cores: 4
[3]:
print(f"Run with ARFS {arfs.__version__}")
Run with ARFS 2.0.5
[4]:
boston = load_data(name="Boston")
X, y = boston.data, boston.target
[5]:
X.dtypes
[5]:
CRIM float64
ZN float64
INDUS float64
CHAS category
NOX float64
RM float64
AGE float64
DIS float64
RAD category
TAX float64
PTRATIO float64
B float64
LSTAT float64
random_num1 float64
random_num2 int32
random_cat category
random_cat_2 category
genuine_num float64
dtype: object
[6]:
X.head()
[6]:
| CRIM | ZN | INDUS | CHAS | NOX | RM | AGE | DIS | RAD | TAX | PTRATIO | B | LSTAT | random_num1 | random_num2 | random_cat | random_cat_2 | genuine_num | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0.00632 | 18.0 | 2.31 | 0.0 | 0.538 | 6.575 | 65.2 | 4.0900 | 1.0 | 296.0 | 15.3 | 396.90 | 4.98 | 0.496714 | 0 | cat_3517 | Platist | 7.080332 |
| 1 | 0.02731 | 0.0 | 7.07 | 0.0 | 0.469 | 6.421 | 78.9 | 4.9671 | 2.0 | 242.0 | 17.8 | 396.90 | 9.14 | -0.138264 | 0 | cat_2397 | MarkZ | 5.245384 |
| 2 | 0.02729 | 0.0 | 7.07 | 0.0 | 0.469 | 7.185 | 61.1 | 4.9671 | 2.0 | 242.0 | 17.8 | 392.83 | 4.03 | 0.647689 | 0 | cat_3735 | Dracula | 6.375795 |
| 3 | 0.03237 | 0.0 | 2.18 | 0.0 | 0.458 | 6.998 | 45.8 | 6.0622 | 3.0 | 222.0 | 18.7 | 394.63 | 2.94 | 1.523030 | 0 | cat_2870 | Bejita | 6.725118 |
| 4 | 0.06905 | 0.0 | 2.18 | 0.0 | 0.458 | 7.147 | 54.2 | 6.0622 | 3.0 | 222.0 | 18.7 | 396.90 | 5.33 | -0.234153 | 4 | cat_1160 | Variance | 7.867781 |
Number of processes#
For optimal performance on CPUs, set the n_jobs parameter to match the number of physical cores available on your machine. Keep in mind that using n_jobs=0 will use the default number of threads in OpenMP, and running multiple processes might not always yield faster results due to the overhead involved in initiating those processes.
[7]:
for n_jobs in range(num_physical_cores):
start_time = time.time()
feat_selector = GrootCV(
objective="rmse",
cutoff=1,
n_folds=5,
n_iter=5,
silent=True,
fastshap=False,
n_jobs=n_jobs,
)
feat_selector.fit(X, y, sample_weight=None)
end_time = time.time()
execution_time = end_time - start_time
print(f"n_jobs = {n_jobs}, Execution time: {execution_time:.3f} seconds")
invalid value encountered in cast
invalid value encountered in cast
invalid value encountered in cast
invalid value encountered in cast
n_jobs = 0, Execution time: 9.602 seconds
n_jobs = 1, Execution time: 8.959 seconds
n_jobs = 2, Execution time: 7.147 seconds
n_jobs = 3, Execution time: 9.987 seconds
Regularization#
By default, GrootCV is using crossvalidation and early-stopping to prevent overfitting and to ensure the robustness of the feature selection. However, if you want to prevent overfitting even more, you can use the dedicated lightgbm parameters such as min_data_in_leaf.
Let’s illustrate: - less regularization than default - default regularization ({"min_data_in_leaf": 20}) - larger regularization
[8]:
# GrootCV with less regularization
feat_selector = GrootCV(
objective="rmse",
cutoff=1,
n_folds=5,
n_iter=5,
silent=True,
fastshap=True,
n_jobs=0,
lgbm_params={"min_data_in_leaf": 10},
)
feat_selector.fit(X, y, sample_weight=None)
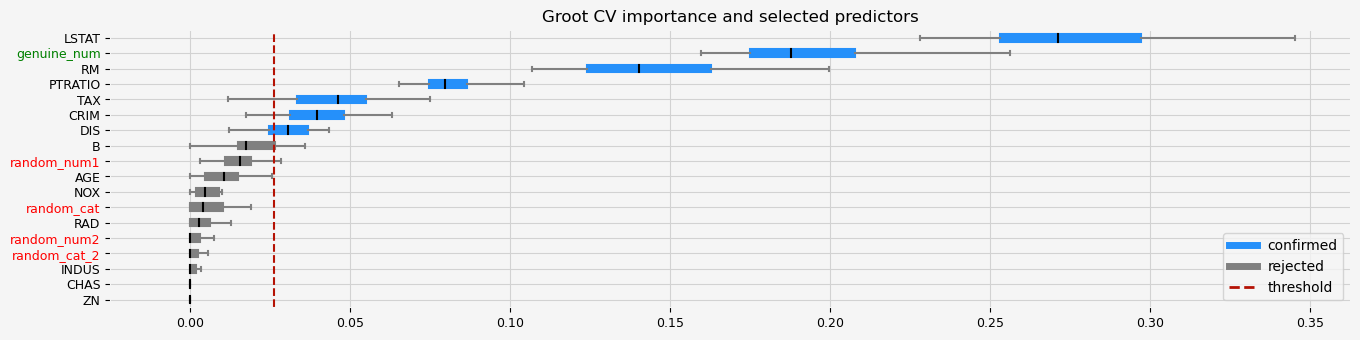
print(f"The selected features: {feat_selector.get_feature_names_out()}")
print(f"The agnostic ranking: {feat_selector.ranking_}")
print(f"The naive ranking: {feat_selector.ranking_absolutes_}")
fig = feat_selector.plot_importance(n_feat_per_inch=5)
# highlight synthetic random variable
fig = highlight_tick(figure=fig, str_match="random")
fig = highlight_tick(figure=fig, str_match="genuine", color="green")
plt.show()
The selected features: ['CRIM' 'NOX' 'RM' 'AGE' 'DIS' 'TAX' 'PTRATIO' 'B' 'LSTAT' 'genuine_num']
The agnostic ranking: [2 1 1 1 2 2 2 2 1 2 2 2 2 1 1 1 1 2]
The naive ranking: ['LSTAT', 'RM', 'genuine_num', 'PTRATIO', 'DIS', 'NOX', 'CRIM', 'AGE', 'TAX', 'B', 'random_num1', 'INDUS', 'random_cat', 'random_cat_2', 'RAD', 'random_num2', 'ZN', 'CHAS']
[9]:
# GrootCV with default regularization
feat_selector = GrootCV(
objective="rmse",
cutoff=1,
n_folds=5,
n_iter=5,
silent=True,
fastshap=True,
n_jobs=0,
lgbm_params=None,
)
feat_selector.fit(X, y, sample_weight=None)
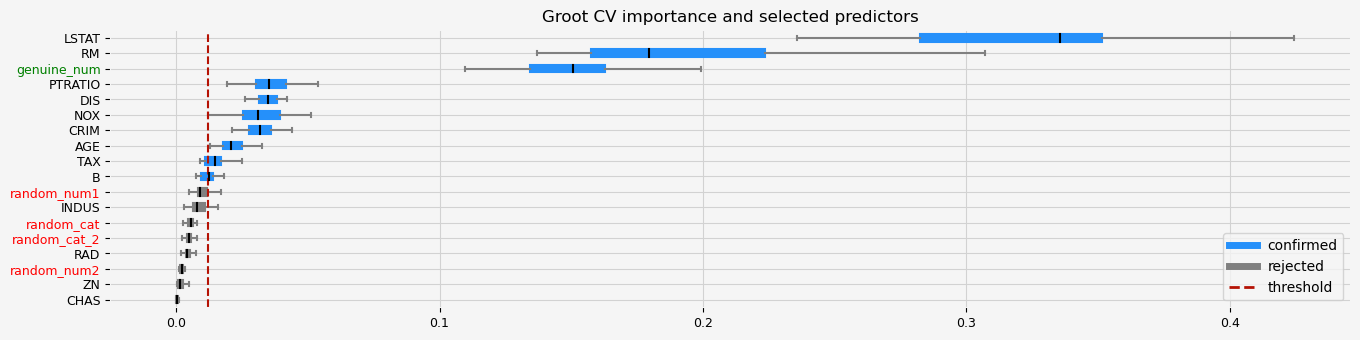
print(f"The selected features: {feat_selector.get_feature_names_out()}")
print(f"The agnostic ranking: {feat_selector.ranking_}")
print(f"The naive ranking: {feat_selector.ranking_absolutes_}")
fig = feat_selector.plot_importance(n_feat_per_inch=5)
# highlight synthetic random variable
fig = highlight_tick(figure=fig, str_match="random")
fig = highlight_tick(figure=fig, str_match="genuine", color="green")
plt.show()
The selected features: ['CRIM' 'NOX' 'RM' 'AGE' 'DIS' 'TAX' 'PTRATIO' 'LSTAT' 'genuine_num']
The agnostic ranking: [2 1 1 1 2 2 2 2 1 2 2 1 2 1 1 1 1 2]
The naive ranking: ['LSTAT', 'RM', 'genuine_num', 'PTRATIO', 'DIS', 'NOX', 'CRIM', 'AGE', 'TAX', 'B', 'random_num1', 'INDUS', 'random_cat', 'random_cat_2', 'RAD', 'ZN', 'random_num2', 'CHAS']
[10]:
# GrootCV with larger regularization
feat_selector = GrootCV(
objective="rmse",
cutoff=1,
n_folds=5,
n_iter=5,
silent=True,
fastshap=True,
n_jobs=0,
lgbm_params={"min_data_in_leaf": 100},
)
feat_selector.fit(X, y, sample_weight=None)
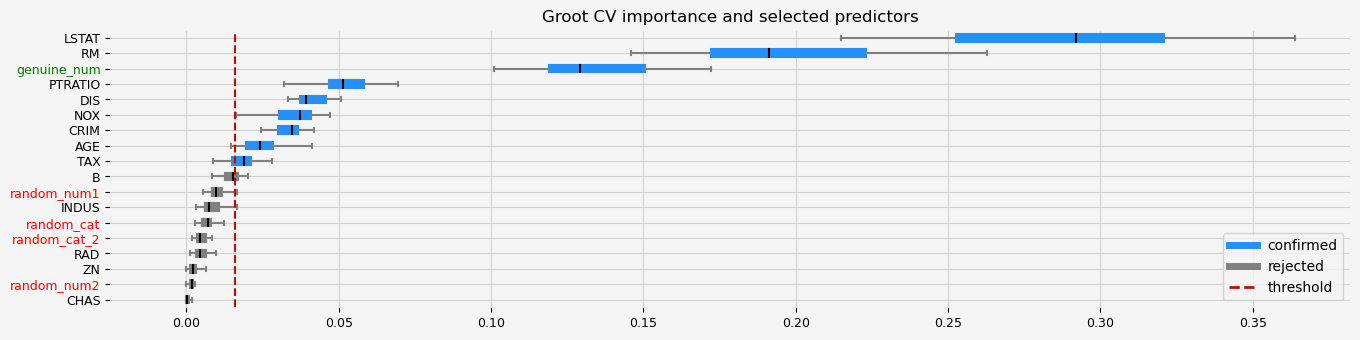
print(f"The selected features: {feat_selector.get_feature_names_out()}")
print(f"The agnostic ranking: {feat_selector.ranking_}")
print(f"The naive ranking: {feat_selector.ranking_absolutes_}")
fig = feat_selector.plot_importance(n_feat_per_inch=5)
# highlight synthetic random variable
fig = highlight_tick(figure=fig, str_match="random")
fig = highlight_tick(figure=fig, str_match="genuine", color="green")
plt.show()
The selected features: ['CRIM' 'RM' 'DIS' 'TAX' 'PTRATIO' 'LSTAT' 'genuine_num']
The agnostic ranking: [2 1 1 1 1 2 1 2 1 2 2 1 2 1 1 1 1 2]
The naive ranking: ['LSTAT', 'genuine_num', 'RM', 'PTRATIO', 'TAX', 'CRIM', 'DIS', 'B', 'random_num1', 'AGE', 'NOX', 'random_cat', 'RAD', 'random_num2', 'random_cat_2', 'INDUS', 'ZN', 'CHAS']