Preprocessing - encoding, discretization, dtypes#
Before performing All Relevant Feature Selection, you might want to apply preprocessing as:
ordinal encoding (recommended over other encoding strategies)
discretization and auto-grouping of levels
select columns in a consistent fashion, according data types and patterns
Ordinal encoding is strongly recommended for GBM-based methods. It is well documented that GBMs generally return better results when categorical predictors are encoded as integers (see lightGBM documentation for instance). OHE should be avoided since it leads to shallow trees.
Auto-grouping levels of categorical predictors and discretization of continuous ones is achieved using a method using GBMs, specifically lightGBM. LightGBM is extremely efficient at building histograms and finding the best splits. Leveraging the inner working of lightGBM, it is fairly simple and blazing fast to discretize and auto-group the predictor matrix.
[1]:
# from IPython.core.display import display, HTML
# display(HTML("<style>.container { width:95% !important; }</style>"))
[2]:
# Settings and libraries
from __future__ import print_function
import pandas as pd
import lightgbm
import numpy as np
import seaborn as sns
from matplotlib import pyplot as plt
import gc
import arfs
import arfs.preprocessing as arfspp
from arfs.utils import (
_make_corr_dataset_regression,
_make_corr_dataset_classification,
)
%matplotlib inline
[3]:
print(f"Run with ARFS {arfs.__version__} and ligthgbm {lightgbm.__version__}")
Run with ARFS 2.1.0 and ligthgbm 3.3.1
Generating artificial data#
Inspired by the BorutaPy unit test
[4]:
X, y, w = _make_corr_dataset_regression()
data = X.copy()
data["target"] = y
[5]:
# significant regressors
x_vars = ["var0", "var1", "var2", "var3", "var4"]
y_vars = ["target"]
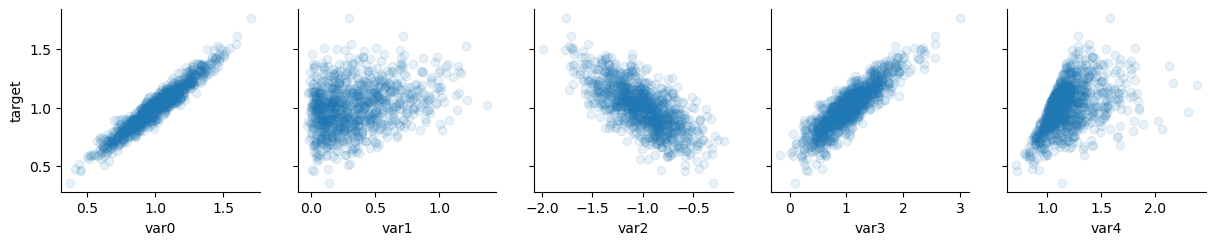
g = sns.PairGrid(data, x_vars=x_vars, y_vars=y_vars)
g.map(plt.scatter, alpha=0.1)
# noise
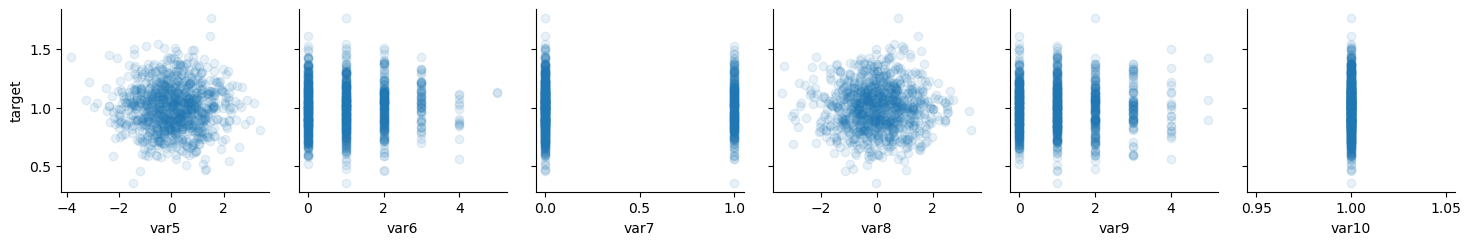
x_vars = ["var5", "var6", "var7", "var8", "var9", "var10"]
y_vars = ["target"]
g = sns.PairGrid(data, x_vars=x_vars, y_vars=y_vars)
g.map(plt.scatter, alpha=0.1)
plt.plot();
[6]:
X.head()
[6]:
| var0 | var1 | var2 | var3 | var4 | var5 | var6 | var7 | var8 | var9 | var10 | var11 | var12 | nice_guys | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1.016361 | 0.821664 | -1.184095 | 0.985738 | 1.050445 | -0.494190 | 2 | 0 | -0.361717 | 1 | 1.0 | 0 | 1.705980 | Bias |
| 1 | 0.929581 | 0.334013 | -1.063030 | 0.819273 | 1.016252 | 0.283845 | 1 | 0 | 0.178670 | 2 | 1.0 | 1 | NaN | Klaue |
| 2 | 1.095456 | 0.187234 | -1.488666 | 1.087443 | 1.140542 | 0.962503 | 2 | 0 | -3.375579 | 2 | 1.0 | 2 | NaN | Imaginedragons |
| 3 | 1.318165 | 0.994528 | -1.370624 | 1.592398 | 1.315021 | 1.165595 | 0 | 0 | -0.449650 | 2 | 1.0 | 3 | 2.929707 | MarkZ |
| 4 | 0.849496 | 0.184859 | -0.806604 | 0.865702 | 0.991916 | -0.058833 | 1 | 0 | 0.763903 | 1 | 1.0 | 4 | NaN | Thanos |
[7]:
X.dtypes
[7]:
var0 float64
var1 float64
var2 float64
var3 float64
var4 float64
var5 float64
var6 int32
var7 int32
var8 float64
var9 int32
var10 float64
var11 category
var12 float64
nice_guys object
dtype: object
Selecting columns#
The custome selector returns a callable (function) which will return the list of column names.
[8]:
from arfs.preprocessing import dtype_column_selector
cat_features_selector = dtype_column_selector(
dtype_include=["category", "object", "bool"],
dtype_exclude=[np.number],
pattern=None,
exclude_cols=None,
)
cat_features_selector(X)
[8]:
['var11', 'nice_guys']
Encoding categorical data#
Unlike to scikit-learn, you don’t need to build a custom ColumnTransformer. You can just throw in a dataframe and it will return the same dataframe with the relevant columns encoded. You can:
fit
transform
fit_transform
inverse_transform
use it in a Pipeline
as you will do in vanilla scikit-learn.
[9]:
from arfs.preprocessing import OrdinalEncoderPandas
encoder = OrdinalEncoderPandas(exclude_cols=["nice_guys"]).fit(X)
X_encoded = encoder.transform(X)
[10]:
X_encoded.head()
[10]:
| var0 | var1 | var2 | var3 | var4 | var5 | var6 | var7 | var8 | var9 | var10 | var11 | var12 | nice_guys | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1.016361 | 0.821664 | -1.184095 | 0.985738 | 1.050445 | -0.494190 | 2 | 0 | -0.361717 | 1 | 1.0 | 0.0 | 1.705980 | Bias |
| 1 | 0.929581 | 0.334013 | -1.063030 | 0.819273 | 1.016252 | 0.283845 | 1 | 0 | 0.178670 | 2 | 1.0 | 1.0 | NaN | Klaue |
| 2 | 1.095456 | 0.187234 | -1.488666 | 1.087443 | 1.140542 | 0.962503 | 2 | 0 | -3.375579 | 2 | 1.0 | 2.0 | NaN | Imaginedragons |
| 3 | 1.318165 | 0.994528 | -1.370624 | 1.592398 | 1.315021 | 1.165595 | 0 | 0 | -0.449650 | 2 | 1.0 | 3.0 | 2.929707 | MarkZ |
| 4 | 0.849496 | 0.184859 | -0.806604 | 0.865702 | 0.991916 | -0.058833 | 1 | 0 | 0.763903 | 1 | 1.0 | 4.0 | NaN | Thanos |
[11]:
encoder.inverse_transform(X_encoded)
[11]:
| var0 | var1 | var2 | var3 | var4 | var5 | var6 | var7 | var8 | var9 | var10 | var11 | var12 | nice_guys | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1.016361 | 0.821664 | -1.184095 | 0.985738 | 1.050445 | -0.494190 | 2 | 0 | -0.361717 | 1 | 1.0 | 0 | 1.705980 | Bias |
| 1 | 0.929581 | 0.334013 | -1.063030 | 0.819273 | 1.016252 | 0.283845 | 1 | 0 | 0.178670 | 2 | 1.0 | 1 | NaN | Klaue |
| 2 | 1.095456 | 0.187234 | -1.488666 | 1.087443 | 1.140542 | 0.962503 | 2 | 0 | -3.375579 | 2 | 1.0 | 2 | NaN | Imaginedragons |
| 3 | 1.318165 | 0.994528 | -1.370624 | 1.592398 | 1.315021 | 1.165595 | 0 | 0 | -0.449650 | 2 | 1.0 | 3 | 2.929707 | MarkZ |
| 4 | 0.849496 | 0.184859 | -0.806604 | 0.865702 | 0.991916 | -0.058833 | 1 | 0 | 0.763903 | 1 | 1.0 | 4 | NaN | Thanos |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 995 | 0.904073 | 0.009764 | -0.909486 | 0.831147 | 1.041658 | 0.081291 | 0 | 0 | -0.542799 | 3 | 1.0 | 495 | NaN | Dracula |
| 996 | 1.370908 | 1.144173 | -1.129008 | 2.002148 | 1.418488 | 0.484591 | 0 | 1 | -1.932372 | 1 | 1.0 | 496 | 2.587139 | Alien |
| 997 | 1.113773 | 0.206798 | -1.371649 | 1.167287 | 1.079760 | 0.267475 | 0 | 0 | -0.329798 | 0 | 1.0 | 497 | 3.880225 | Gruber |
| 998 | 0.905654 | 0.598419 | -0.792174 | 0.783557 | 1.275266 | 0.380826 | 1 | 0 | -1.987310 | 1 | 1.0 | 498 | 1.888725 | Gruber |
| 999 | 1.138102 | 0.143818 | -1.348573 | 1.321900 | 1.309521 | 1.342722 | 1 | 0 | -0.871590 | 1 | 1.0 | 499 | NaN | Creationist |
1000 rows × 14 columns
[12]:
X.head()
[12]:
| var0 | var1 | var2 | var3 | var4 | var5 | var6 | var7 | var8 | var9 | var10 | var11 | var12 | nice_guys | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1.016361 | 0.821664 | -1.184095 | 0.985738 | 1.050445 | -0.494190 | 2 | 0 | -0.361717 | 1 | 1.0 | 0 | 1.705980 | Bias |
| 1 | 0.929581 | 0.334013 | -1.063030 | 0.819273 | 1.016252 | 0.283845 | 1 | 0 | 0.178670 | 2 | 1.0 | 1 | NaN | Klaue |
| 2 | 1.095456 | 0.187234 | -1.488666 | 1.087443 | 1.140542 | 0.962503 | 2 | 0 | -3.375579 | 2 | 1.0 | 2 | NaN | Imaginedragons |
| 3 | 1.318165 | 0.994528 | -1.370624 | 1.592398 | 1.315021 | 1.165595 | 0 | 0 | -0.449650 | 2 | 1.0 | 3 | 2.929707 | MarkZ |
| 4 | 0.849496 | 0.184859 | -0.806604 | 0.865702 | 0.991916 | -0.058833 | 1 | 0 | 0.763903 | 1 | 1.0 | 4 | NaN | Thanos |
[13]:
X.nunique()
[13]:
var0 1000
var1 1000
var2 1000
var3 1000
var4 1000
var5 1000
var6 6
var7 2
var8 1000
var9 6
var10 1
var11 500
var12 500
nice_guys 36
dtype: int64
Discretization and auto-grouping#
Discretization and auto-grouping are supervised tasks and therefore require a target, and optionally a weight.
[14]:
from arfs.preprocessing import TreeDiscretizer
# main parameter controlling how agressive will be the auto-grouping
lgb_params = {"min_split_gain": 0.05}
# instanciate the discretizer
disc = TreeDiscretizer(bin_features="all", n_bins=10, boost_params=lgb_params)
disc.fit(X=X, y=y, sample_weight=w)
[14]:
TreeDiscretizer(bin_features=['var0', 'var1', 'var2', 'var3', 'var4', 'var5',
'var6', 'var7', 'var8', 'var9', 'var10', 'var11',
'var12', 'nice_guys'],
boost_params={'max_leaf': 10, 'min_split_gain': 0.05,
'num_boost_round': 1, 'objective': 'rmse'})In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook. On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
TreeDiscretizer(bin_features=['var0', 'var1', 'var2', 'var3', 'var4', 'var5',
'var6', 'var7', 'var8', 'var9', 'var10', 'var11',
'var12', 'nice_guys'],
boost_params={'max_leaf': 10, 'min_split_gain': 0.05,
'num_boost_round': 1, 'objective': 'rmse'})The regularization min_split_gain has as an outcome, as expected, to auto-group into a single level all the predictors not correlated to the target.
[15]:
X_trans = disc.transform(X)
X_trans
[15]:
| var0 | var1 | var2 | var3 | var4 | var5 | var6 | var7 | var8 | var9 | var10 | var11 | var12 | nice_guys | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | (1.01, 1.09] | (0.6, 0.85] | (-1.34, -1.1] | (0.84, 1.01] | (1.0, 1.06] | (-0.96, -0.24] | (1.0, inf] | (-inf, inf] | (-1.43, -0.00048] | (-inf, inf] | (-inf, inf] | 0 / 1 //.../10 / 11 | (1.61, 2.55] | Bias / /...// Excel |
| 1 | (0.89, 1.01] | (0.32, 0.35] | (-1.1, -0.96] | (0.71, 0.84] | (1.0, 1.06] | (0.084, 0.34] | (-inf, 1.0] | (-inf, inf] | (-0.00048, 0.18] | (-inf, inf] | (-inf, inf] | 0 / 1 //.../10 / 11 | (1.61, 2.55] | Bias / /...// Excel |
| 2 | (1.09, 1.22] | (0.17, 0.27] | (-1.58, -1.34] | (1.01, 1.12] | (1.13, 1.34] | (0.68, 1.45] | (1.0, inf] | (-inf, inf] | (-inf, -1.91] | (-inf, inf] | (-inf, inf] | 0 / 1 //.../10 / 11 | (1.61, 2.55] | Bias / /...// Excel |
| 3 | (1.3, 1.38] | (0.85, inf] | (-1.58, -1.34] | (1.42, 1.6] | (1.13, 1.34] | (0.68, 1.45] | (-inf, 1.0] | (-inf, inf] | (-1.43, -0.00048] | (-inf, inf] | (-inf, inf] | 0 / 1 //.../10 / 11 | (2.88, inf] | Bias / /...// Excel |
| 4 | (0.78, 0.89] | (0.17, 0.27] | (-0.89, -0.59] | (0.84, 1.01] | (0.9, 1.0] | (-0.12, 0.084] | (-inf, 1.0] | (-inf, inf] | (0.72, 0.98] | (-inf, inf] | (-inf, inf] | 0 / 1 //.../10 / 11 | (1.61, 2.55] | Bias / /...// Excel |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 995 | (0.89, 1.01] | (-inf, 0.17] | (-0.96, -0.89] | (0.71, 0.84] | (1.0, 1.06] | (-0.12, 0.084] | (-inf, 1.0] | (-inf, inf] | (-1.43, -0.00048] | (-inf, inf] | (-inf, inf] | 468 / 4/.../8 / 499 | (1.61, 2.55] | Bias / /...// Excel |
| 996 | (1.3, 1.38] | (0.85, inf] | (-1.34, -1.1] | (1.81, inf] | (1.42, inf] | (0.44, 0.63] | (-inf, 1.0] | (-inf, inf] | (-inf, -1.91] | (-inf, inf] | (-inf, inf] | 468 / 4/.../8 / 499 | (2.55, 2.88] | Bias / /...// Excel |
| 997 | (1.09, 1.22] | (0.17, 0.27] | (-1.58, -1.34] | (1.12, 1.31] | (1.06, 1.13] | (0.084, 0.34] | (-inf, 1.0] | (-inf, inf] | (-1.43, -0.00048] | (-inf, inf] | (-inf, inf] | 468 / 4/.../8 / 499 | (2.88, inf] | Bias / /...// Excel |
| 998 | (0.89, 1.01] | (0.45, 0.6] | (-0.89, -0.59] | (0.71, 0.84] | (1.13, 1.34] | (0.34, 0.44] | (-inf, 1.0] | (-inf, inf] | (-inf, -1.91] | (-inf, inf] | (-inf, inf] | 468 / 4/.../8 / 499 | (1.61, 2.55] | Bias / /...// Excel |
| 999 | (1.09, 1.22] | (-inf, 0.17] | (-1.58, -1.34] | (1.31, 1.42] | (1.13, 1.34] | (0.68, 1.45] | (-inf, 1.0] | (-inf, inf] | (-1.43, -0.00048] | (-inf, inf] | (-inf, inf] | 468 / 4/.../8 / 499 | (1.61, 2.55] | Bias / /...// Excel |
1000 rows × 14 columns
Sklearn Pipeline#
The selectors follow the scikit-learn base class, therefore they are compatible with scikit-learn in general. This has several advantages: - easier to maintain - easier to version - more flexible - running faster by removing unnecessary columns before going to the computational demanding steps
[16]:
from sklearn.pipeline import Pipeline
from arfs.preprocessing import OrdinalEncoderPandas
from arfs.feature_selection import CollinearityThreshold, UniqueValuesThreshold, MissingValueThreshold
encoder = Pipeline(
[ ("zero_variance", UniqueValuesThreshold()),
("collinearity", CollinearityThreshold(threshold=0.75)),
("disctretizer", TreeDiscretizer(bin_features="all", n_bins=10, boost_params=lgb_params)),
# the treediscretization might introduce NaN for pure noise columns
("zero_variance_disc", UniqueValuesThreshold()),
("encoder", OrdinalEncoderPandas()),
]
)
X_encoded = encoder.fit(X, y).transform(X)
X_encoded.head()
[16]:
| var1 | var3 | var5 | var6 | var8 | |
|---|---|---|---|---|---|
| 0 | (0.74, 0.94] | (0.91, 1.12] | (-inf, -0.31] | (1.0, inf] | (-0.45, -0.28] |
| 1 | (0.19, 0.4] | (0.71, 0.91] | (0.084, 0.34] | (-inf, 1.0] | (-0.00048, 0.18] |
| 2 | (0.16, 0.19] | (0.91, 1.12] | (0.45, 1.03] | (1.0, inf] | (-inf, -1.15] |
| 3 | (0.94, inf] | (1.48, 1.6] | (1.03, 1.38] | (-inf, 1.0] | (-0.59, -0.45] |
| 4 | (0.16, 0.19] | (0.71, 0.91] | (-0.17, 0.084] | (-inf, 1.0] | (0.33, 0.98] |