ARFS - classification#
ARFS can be used for classification (binary or multi-class) and for regression. You just have to specify the right loss function.
[1]:
# from IPython.core.display import display, HTML
# display(HTML("<style>.container { width:95% !important; }</style>"))
import catboost
import numpy as np
import pandas as pd
import matplotlib as mpl
import matplotlib.pyplot as plt
import gc
import shap
from boruta import BorutaPy as bp
from sklearn.datasets import fetch_openml
from sklearn.inspection import permutation_importance
from sklearn.pipeline import Pipeline
from sklearn.datasets import fetch_openml
from sklearn.inspection import permutation_importance
from sklearn.base import clone
from sklearn.ensemble import RandomForestRegressor, RandomForestClassifier
from lightgbm import LGBMRegressor, LGBMClassifier
from xgboost import XGBRegressor, XGBClassifier
from catboost import CatBoostRegressor, CatBoostClassifier
from sys import getsizeof, path
import arfs
import arfs.feature_selection as arfsfs
import arfs.feature_selection.allrelevant as arfsgroot
from arfs.feature_selection import (
MinRedundancyMaxRelevance,
GrootCV,
MissingValueThreshold,
UniqueValuesThreshold,
CollinearityThreshold,
make_fs_summary,
)
from arfs.utils import LightForestClassifier, LightForestRegressor
from arfs.benchmark import highlight_tick, compare_varimp, sklearn_pimp_bench
from arfs.utils import load_data
plt.style.use("fivethirtyeight")
rng = np.random.RandomState(seed=42)
# import warnings
# warnings.filterwarnings('ignore')
Using `tqdm.autonotebook.tqdm` in notebook mode. Use `tqdm.tqdm` instead to force console mode (e.g. in jupyter console)
[2]:
print(f"Run with ARFS {arfs.__version__}")
Run with ARFS 2.0.5
[3]:
import importlib
importlib.reload(arfs)
[3]:
<module 'arfs' from '/home/bsatom/Documents/arfs/src/arfs/__init__.py'>
[4]:
%matplotlib inline
[5]:
gc.enable()
gc.collect()
[5]:
4
Simple Usage#
In the following examples, I’ll use a classical data set to which I added random predictors (numerical and categorical) and a genuine but noisy artificial predictor (correlated to the target). An All Relveant FS methods should discard them. In the unit tests, you’ll find examples using artifical data with genuine (correlated and non-linear) predictors and with some random/noise columns.
[6]:
# titanic = load_data(name='Titanic')
# X, y = titanic.data, titanic.target
[7]:
titanic = load_data(name="Titanic")
X, y = titanic.data, titanic.target
y = y.astype(int)
The default value of `parser` will change from `'liac-arff'` to `'auto'` in 1.4. You can set `parser='auto'` to silence this warning. Therefore, an `ImportError` will be raised from 1.4 if the dataset is dense and pandas is not installed. Note that the pandas parser may return different data types. See the Notes Section in fetch_openml's API doc for details.
Leshy#
[8]:
# Let's use lightgbm as booster, see below for using more models
model = LGBMClassifier(random_state=42, verbose=-1)
# model = CatBoostClassifier(random_state=42, verbose=0)
# model = XGBClassifier(random_state=42, verbosity=0)
with native (impurity/Gini) importance
[9]:
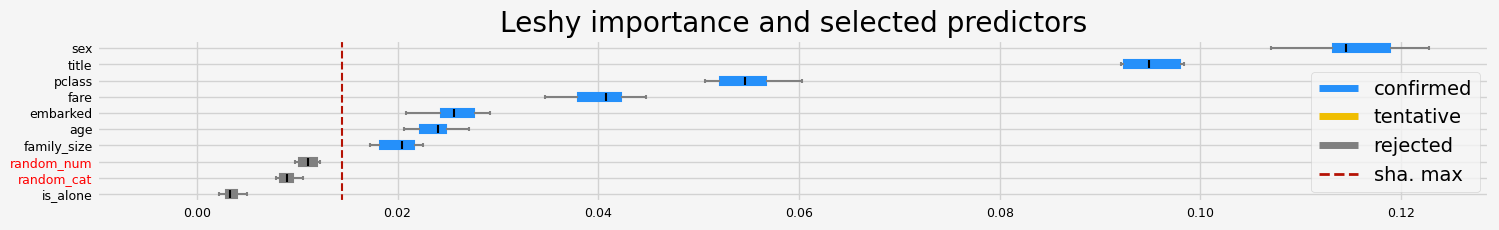
%%time
# Leshy
feat_selector = arfsgroot.Leshy(
model, n_estimators=20, verbose=1, max_iter=10, random_state=42, importance="native"
)
feat_selector.fit(X, y, sample_weight=None)
print(f"The selected features: {feat_selector.get_feature_names_out()}")
print(f"The agnostic ranking: {feat_selector.ranking_}")
print(f"The naive ranking: {feat_selector.ranking_absolutes_}")
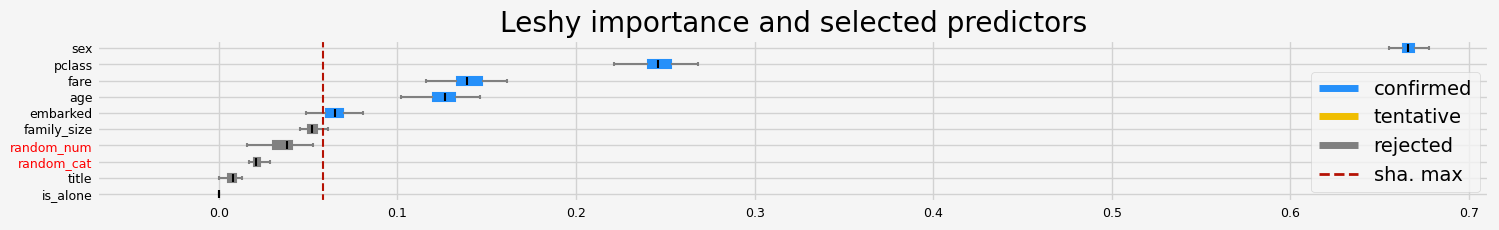
fig = feat_selector.plot_importance(n_feat_per_inch=5)
# highlight synthetic random variable
fig = highlight_tick(figure=fig, str_match="random")
fig = highlight_tick(figure=fig, str_match="genuine", color="green")
plt.show()
Leshy finished running using native var. imp.
Iteration: 1 / 10
Confirmed: 2
Tentative: 0
Rejected: 8
All relevant predictors selected in 00:00:00.55
The selected features: ['age' 'fare']
The agnostic ranking: [3 6 6 4 9 8 1 5 1 2]
The naive ranking: ['fare', 'age', 'random_num', 'pclass', 'random_cat', 'family_size', 'embarked', 'sex', 'title', 'is_alone']
CPU times: user 1.53 s, sys: 160 ms, total: 1.69 s
Wall time: 1.07 s
with SHAP importance, original implementation
[10]:
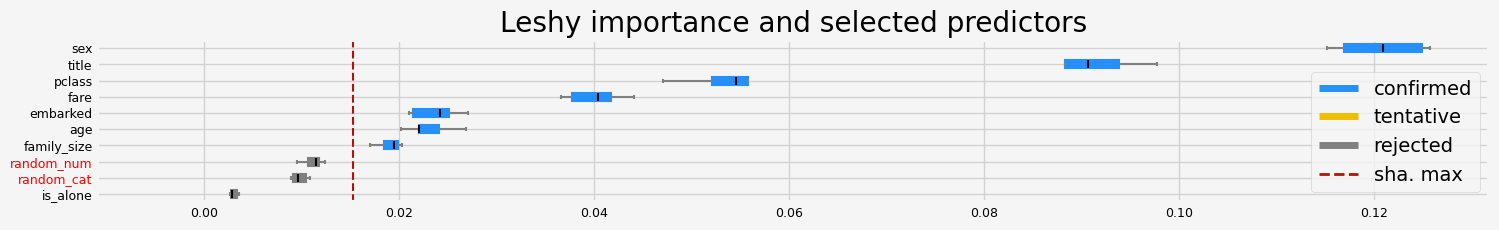
%%time
# Leshy
model = clone(model)
feat_selector = arfsgroot.Leshy(
model, n_estimators=50, verbose=1, max_iter=10, random_state=42, importance="shap"
)
feat_selector.fit(X, y, sample_weight=None)
print(f"The selected features: {feat_selector.get_feature_names_out()}")
print(f"The agnostic ranking: {feat_selector.ranking_}")
print(f"The naive ranking: {feat_selector.ranking_absolutes_}")
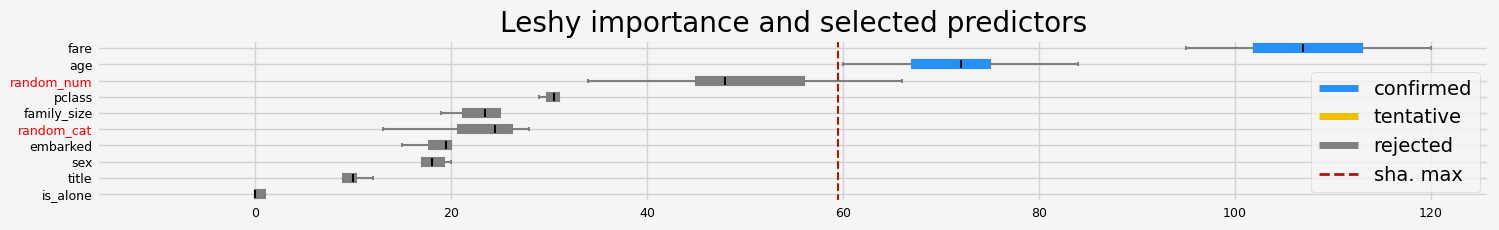
fig = feat_selector.plot_importance(n_feat_per_inch=5)
# highlight synthetic random variable
fig = highlight_tick(figure=fig, str_match="random")
fig = highlight_tick(figure=fig, str_match="genuine", color="green")
plt.show()
Leshy finished running using shap var. imp.
Iteration: 1 / 10
Confirmed: 4
Tentative: 2
Rejected: 4
All relevant predictors selected in 00:00:01.80
The selected features: ['pclass' 'sex' 'age' 'fare']
The agnostic ranking: [1 1 2 4 6 5 1 2 1 3]
The naive ranking: ['sex', 'pclass', 'fare', 'age', 'embarked', 'family_size', 'random_num', 'random_cat', 'title', 'is_alone']
CPU times: user 3.66 s, sys: 246 ms, total: 3.91 s
Wall time: 2.21 s
the LinkedIn fasttreeshap implementation
[11]:
%%time
# Leshy
model = clone(model)
feat_selector = arfsgroot.Leshy(
model,
n_estimators=50,
verbose=1,
max_iter=10,
random_state=42,
importance="fastshap",
)
feat_selector.fit(X, y, sample_weight=None)
print(f"The selected features: {feat_selector.get_feature_names_out()}")
print(f"The agnostic ranking: {feat_selector.ranking_}")
print(f"The naive ranking: {feat_selector.ranking_absolutes_}")
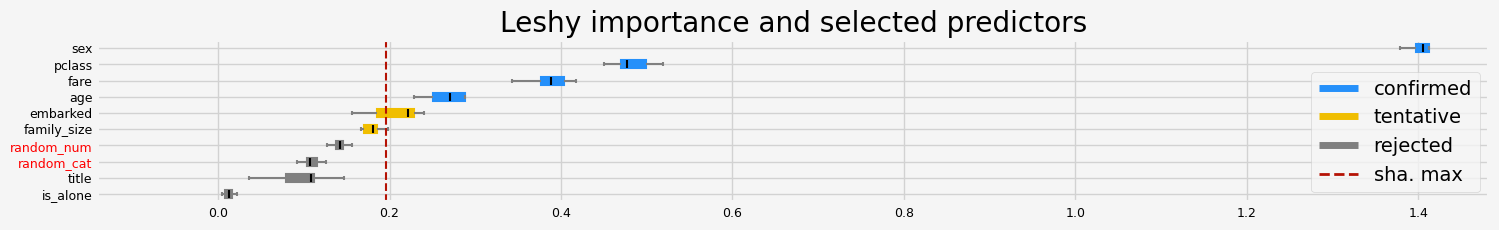
fig = feat_selector.plot_importance(n_feat_per_inch=5)
# highlight synthetic random variable
fig = highlight_tick(figure=fig, str_match="random")
fig = highlight_tick(figure=fig, str_match="genuine", color="green")
plt.show()
Leshy finished running using native var. imp.
Iteration: 1 / 10
Confirmed: 4
Tentative: 2
Rejected: 4
All relevant predictors selected in 00:00:01.88
The selected features: ['pclass' 'sex' 'age' 'fare']
The agnostic ranking: [1 1 2 4 6 5 1 2 1 3]
The naive ranking: ['sex', 'pclass', 'fare', 'age', 'embarked', 'family_size', 'random_num', 'random_cat', 'title', 'is_alone']
CPU times: user 4.55 s, sys: 227 ms, total: 4.77 s
Wall time: 2.25 s
with permutation importance
[12]:
%%time
# Leshy
model = clone(model)
feat_selector = arfsgroot.Leshy(
model, n_estimators=50, verbose=1, max_iter=10, random_state=42, importance="pimp"
)
feat_selector.fit(X, y, sample_weight=None)
print(f"The selected features: {feat_selector.get_feature_names_out()}")
print(f"The agnostic ranking: {feat_selector.ranking_}")
print(f"The naive ranking: {feat_selector.ranking_absolutes_}")
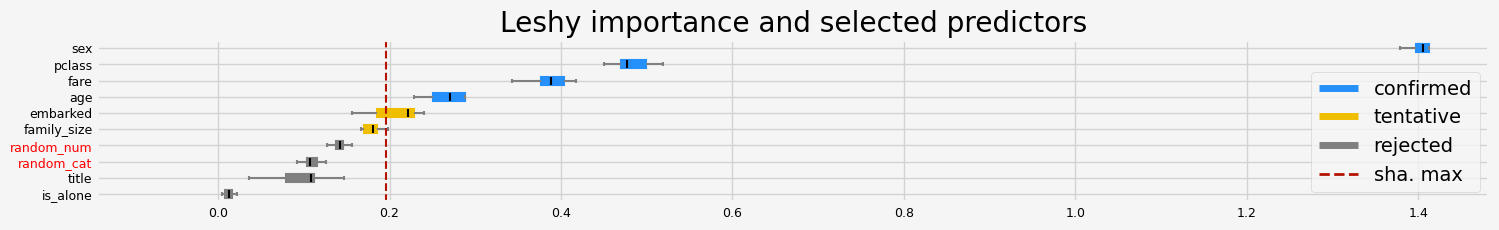
fig = feat_selector.plot_importance(n_feat_per_inch=5)
# highlight synthetic random variable
fig = highlight_tick(figure=fig, str_match="random")
fig = highlight_tick(figure=fig, str_match="genuine", color="green")
plt.show()
Leshy finished running using pimp var. imp.
Iteration: 1 / 10
Confirmed: 4
Tentative: 2
Rejected: 4
All relevant predictors selected in 00:00:07.64
The selected features: ['pclass' 'sex' 'age' 'family_size']
The agnostic ranking: [1 1 2 6 4 3 1 1 2 5]
The naive ranking: ['sex', 'pclass', 'age', 'family_size', 'fare', 'embarked', 'title', 'is_alone', 'random_num', 'random_cat']

CPU times: user 3.03 s, sys: 442 ms, total: 3.47 s
Wall time: 8.07 s
BoostAGroota#
with permutation importance
[13]:
%%time
# be sure to use the same but non-fitted estimator
model = clone(model)
# BoostAGroota
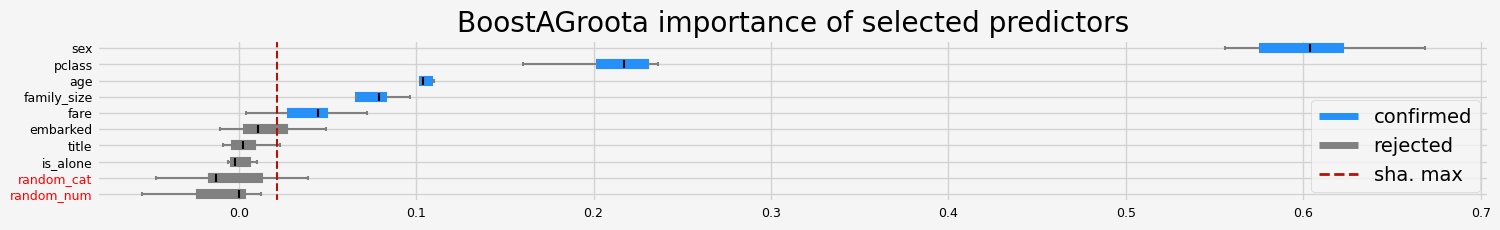
feat_selector = arfsgroot.BoostAGroota(
estimator=model, cutoff=1, iters=10, max_rounds=10, delta=0.1, importance="pimp"
)
feat_selector.fit(X, y, sample_weight=None)
print(f"The selected features: {feat_selector.get_feature_names_out()}")
print(f"The agnostic ranking: {feat_selector.ranking_}")
print(f"The naive ranking: {feat_selector.ranking_absolutes_}")
fig = feat_selector.plot_importance(n_feat_per_inch=5)
# highlight synthetic random variable
fig = highlight_tick(figure=fig, str_match="random")
fig = highlight_tick(figure=fig, str_match="genuine", color="green")
plt.show()
The selected features: ['pclass' 'sex' 'age' 'family_size' 'fare']
The agnostic ranking: [2 2 1 1 1 1 2 2 2 1]
The naive ranking: ['sex', 'pclass', 'age', 'family_size', 'fare', 'embarked', 'title', 'is_alone', 'random_cat', 'random_num']
CPU times: user 4.51 s, sys: 394 ms, total: 4.91 s
Wall time: 9.15 s
with SHAP importance
[14]:
%%time
# be sure to use the same but non-fitted estimator
model = clone(model)
# BoostAGroota
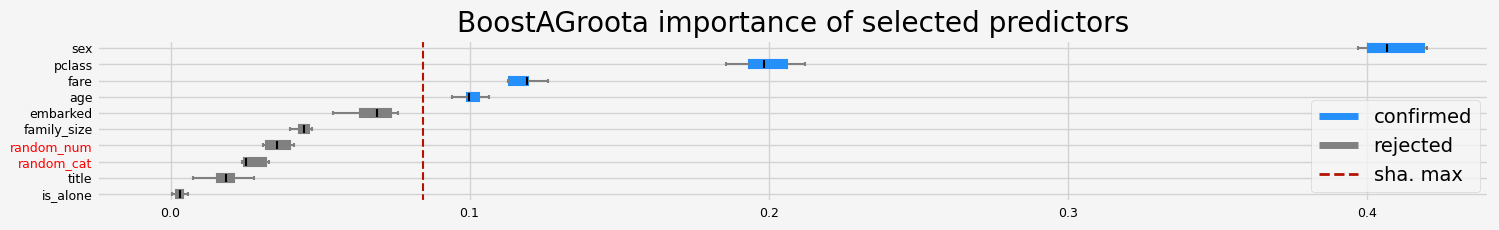
feat_selector = arfsgroot.BoostAGroota(
estimator=model, cutoff=1, iters=10, max_rounds=10, delta=0.1, importance="shap"
)
feat_selector.fit(X, y, sample_weight=None)
print(f"The selected features: {feat_selector.get_feature_names_out()}")
print(f"The agnostic ranking: {feat_selector.ranking_}")
print(f"The naive ranking: {feat_selector.ranking_absolutes_}")
fig = feat_selector.plot_importance(n_feat_per_inch=5)
# highlight synthetic random variable
fig = highlight_tick(figure=fig, str_match="random")
fig = highlight_tick(figure=fig, str_match="genuine", color="green")
plt.show()
The selected features: ['pclass' 'sex' 'age' 'fare']
The agnostic ranking: [2 2 1 1 1 1 2 1 2 1]
The naive ranking: ['sex', 'pclass', 'fare', 'age', 'embarked', 'family_size', 'random_num', 'random_cat', 'title', 'is_alone']
CPU times: user 7.99 s, sys: 339 ms, total: 8.33 s
Wall time: 4.79 s
[15]:
%%time
# be sure to use the same but non-fitted estimator
model = clone(model)
# BoostAGroota
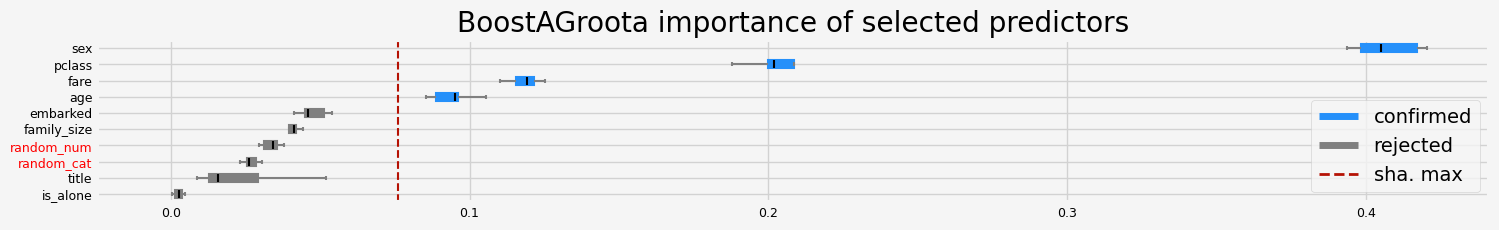
feat_selector = arfsgroot.BoostAGroota(
estimator=model, cutoff=1, iters=10, max_rounds=10, delta=0.1, importance="fastshap"
)
feat_selector.fit(X, y, sample_weight=None)
print(f"The selected features: {feat_selector.get_feature_names_out()}")
print(f"The agnostic ranking: {feat_selector.ranking_}")
print(f"The naive ranking: {feat_selector.ranking_absolutes_}")
fig = feat_selector.plot_importance(n_feat_per_inch=5)
# highlight synthetic random variable
fig = highlight_tick(figure=fig, str_match="random")
fig = highlight_tick(figure=fig, str_match="genuine", color="green")
plt.show()
The selected features: ['pclass' 'sex' 'age' 'fare']
The agnostic ranking: [2 2 1 1 1 1 2 1 2 1]
The naive ranking: ['sex', 'pclass', 'fare', 'age', 'embarked', 'family_size', 'random_num', 'random_cat', 'title', 'is_alone']
CPU times: user 5.98 s, sys: 348 ms, total: 6.33 s
Wall time: 3.77 s
GrootCV#
Internally, it uses lightGBM and SHAP importance (fast and accurate)
[16]:
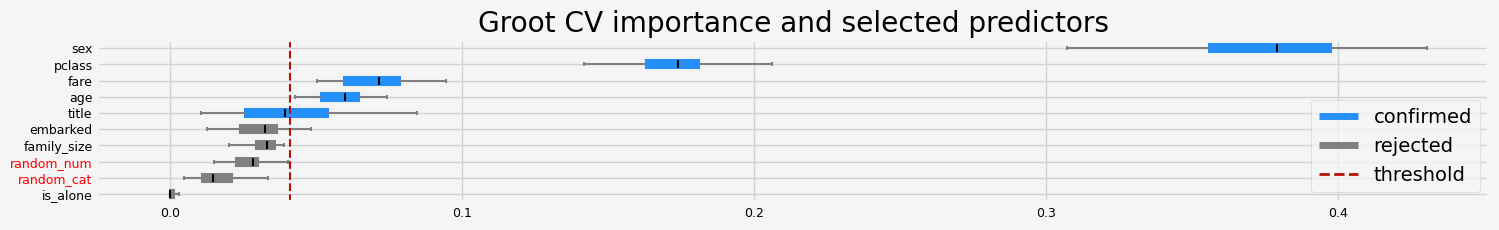
%%time
# GrootCV
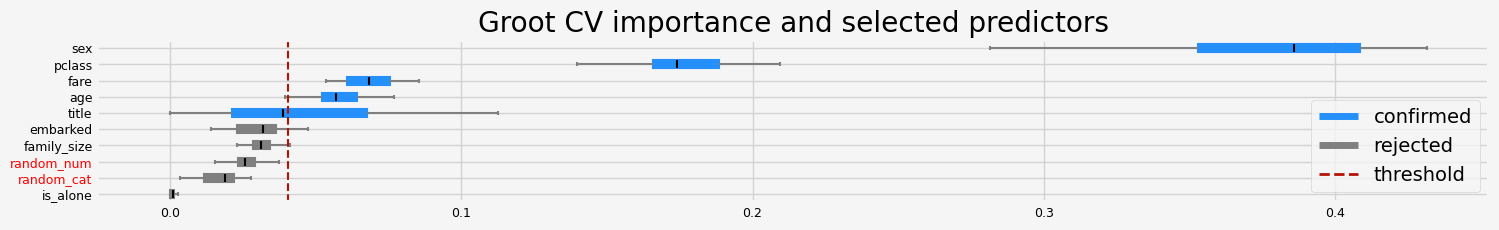
feat_selector = arfsgroot.GrootCV(
objective="binary", cutoff=1, n_folds=5, n_iter=5, silent=True, fastshap=False
)
feat_selector.fit(X, y, sample_weight=None)
print(f"The selected features: {feat_selector.get_feature_names_out()}")
print(f"The agnostic ranking: {feat_selector.ranking_}")
print(f"The naive ranking: {feat_selector.ranking_absolutes_}")
fig = feat_selector.plot_importance(n_feat_per_inch=5)
# highlight synthetic random variable
fig = highlight_tick(figure=fig, str_match="random")
fig = highlight_tick(figure=fig, str_match="genuine", color="green")
plt.show()
The selected features: ['pclass' 'sex' 'title' 'age' 'fare']
The agnostic ranking: [2 2 1 1 1 2 2 1 2 1]
The naive ranking: ['sex', 'pclass', 'fare', 'age', 'title', 'embarked', 'family_size', 'random_num', 'random_cat', 'is_alone']
CPU times: user 14.8 s, sys: 1.14 s, total: 16 s
Wall time: 6.57 s
For larger datasets, the fasttreshap implementation can speed up the feature selection process. However, for smaller datasets, the overhead might slightly slow down the process.
[17]:
%%time
# GrootCV
feat_selector = arfsgroot.GrootCV(
objective="binary", cutoff=1, n_folds=5, n_iter=5, silent=True, fastshap=True
)
feat_selector.fit(X, y, sample_weight=None)
print(f"The selected features: {feat_selector.get_feature_names_out()}")
print(f"The agnostic ranking: {feat_selector.ranking_}")
print(f"The naive ranking: {feat_selector.ranking_absolutes_}")
fig = feat_selector.plot_importance(n_feat_per_inch=5)
# highlight synthetic random variable
fig = highlight_tick(figure=fig, str_match="random")
fig = highlight_tick(figure=fig, str_match="genuine", color="green")
plt.show()
The selected features: ['pclass' 'sex' 'title' 'age' 'fare']
The agnostic ranking: [2 2 1 1 1 2 2 1 2 1]
The naive ranking: ['sex', 'pclass', 'fare', 'age', 'title', 'embarked', 'family_size', 'random_num', 'random_cat', 'is_alone']
CPU times: user 12.8 s, sys: 1.12 s, total: 13.9 s
Wall time: 6.4 s
ARFS in sklearn pipelines#
all the selectors (basic, arfs and MRmr) are sklearn compatible and follows the same architecture. Namely, they use the sklearn relevant base classes and therefore have the same methods.
[18]:
model = clone(model)
# # Leshi/Boruta
# feat_selector = arfsgroot.Leshy(model, n_estimators=50, verbose=1, max_iter=10, random_state=42, importance='shap')
# BoostAGroota
feat_selector = arfsgroot.BoostAGroota(
estimator=model, cutoff=1, iters=10, max_rounds=10, delta=0.1, importance="shap"
)
# GrootCV
# feat_selector = arfsgroot.GrootCV(objective='binary', cutoff=1, n_folds=5, n_iter=5, silent=True)
arfs_fs_pipeline = Pipeline(
[
("missing", MissingValueThreshold(threshold=0.05)),
("unique", UniqueValuesThreshold(threshold=1)),
("collinearity", CollinearityThreshold(threshold=0.85)),
("arfs", feat_selector),
]
)
X_trans = arfs_fs_pipeline.fit(X=X, y=y).transform(X=X)
[19]:
arfs_fs_pipeline.named_steps["collinearity"].get_feature_names_out()
[19]:
array(['pclass', 'embarked', 'random_cat', 'is_alone', 'title',
'random_num'], dtype=object)
[20]:
fig = arfs_fs_pipeline.named_steps["arfs"].plot_importance()
# highlight synthetic random variable
fig = highlight_tick(figure=fig, str_match="random")
fig = highlight_tick(figure=fig, str_match="genuine", color="green")
plt.show()
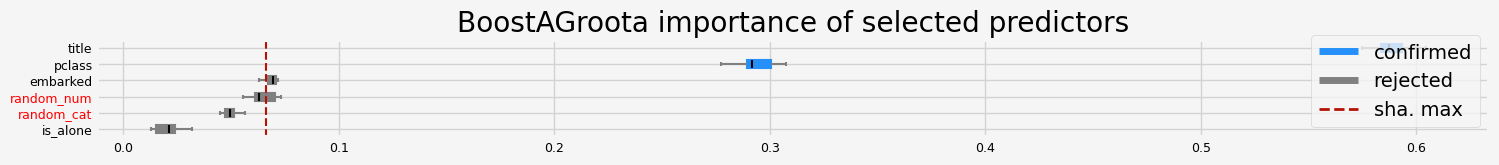
Testing and comparing Leshy, GrootCV and BoostAGroota#
In the following examples, I’ll use different models which are scikit-learn compatible and then one can compare the different ARFS methods with different models and the different feature importance. For Leshy (Boruta) and BoostAGroota, the native feature importance (gini/impurity) returns the less reliable results.
[21]:
%%time
model = clone(model)
# Benchmark with scikit-learn permutation importance
print("=" * 20 + " Benchmarking using sklearn permutation importance " + "=" * 20)
fig = sklearn_pimp_bench(model, X, y, task="classification", sample_weight=None)
==================== Benchmarking using sklearn permutation importance ====================

CPU times: user 777 ms, sys: 336 ms, total: 1.11 s
Wall time: 3.1 s
[22]:
model.__class__.__name__
[22]:
'LGBMClassifier'
Testing Leshy#
[23]:
models = [
RandomForestClassifier(n_jobs=4, oob_score=True),
CatBoostClassifier(random_state=42, verbose=0),
LGBMClassifier(random_state=42, verbose=-1),
LightForestClassifier(n_feat=X.shape[1]),
XGBClassifier(random_state=42, verbosity=0, eval_metric="logloss"),
]
feat_selector = arfsgroot.Leshy(
model, n_estimators=100, verbose=1, max_iter=10, random_state=42
)
if __name__ == "__main__":
# classification
titanic = load_data(name="Titanic")
X, y = titanic.data, titanic.target.cat.codes
cat_f = titanic.categorical
# running the ARFS methods using different models
compare_varimp(feat_selector, models, X, y, sample_weight=None)
==================== Leshy - testing: RandomForestClassifier for var.imp: shap ====================
The default value of `parser` will change from `'liac-arff'` to `'auto'` in 1.4. You can set `parser='auto'` to silence this warning. Therefore, an `ImportError` will be raised from 1.4 if the dataset is dense and pandas is not installed. Note that the pandas parser may return different data types. See the Notes Section in fetch_openml's API doc for details.
Leshy finished running using shap var. imp.
Iteration: 1 / 10
Confirmed: 7
Tentative: 0
Rejected: 3
All relevant predictors selected in 00:00:31.23
['pclass' 'sex' 'embarked' 'title' 'age' 'family_size' 'fare']
==================== Leshy - testing: RandomForestClassifier for var.imp: fastshap ====================
Leshy finished running using native var. imp.
Iteration: 1 / 10
Confirmed: 7
Tentative: 0
Rejected: 3
All relevant predictors selected in 00:00:11.42
['pclass' 'sex' 'embarked' 'title' 'age' 'family_size' 'fare']
==================== Leshy - testing: RandomForestClassifier for var.imp: pimp ====================
Leshy finished running using pimp var. imp.
Iteration: 1 / 10
Confirmed: 5
Tentative: 1
Rejected: 4
All relevant predictors selected in 00:00:22.61
['pclass' 'sex' 'title' 'age' 'family_size']
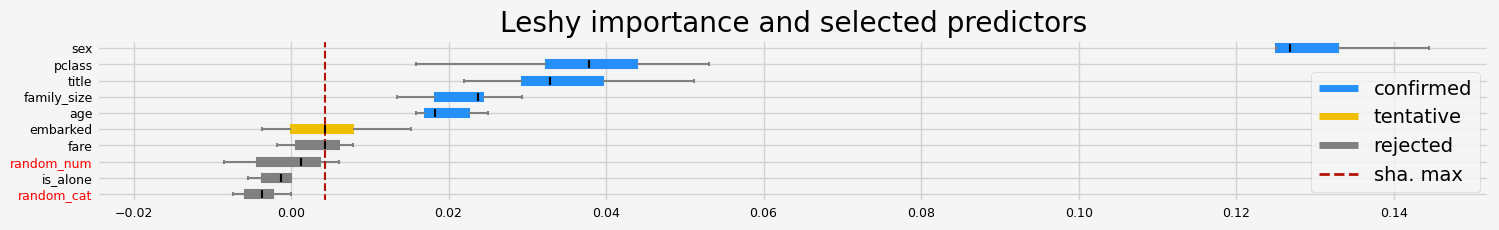
==================== Leshy - testing: RandomForestClassifier for var.imp: native ====================
Leshy finished running using native var. imp.
Iteration: 1 / 10
Confirmed: 4
Tentative: 1
Rejected: 5
All relevant predictors selected in 00:00:04.49
['sex' 'title' 'age' 'fare']
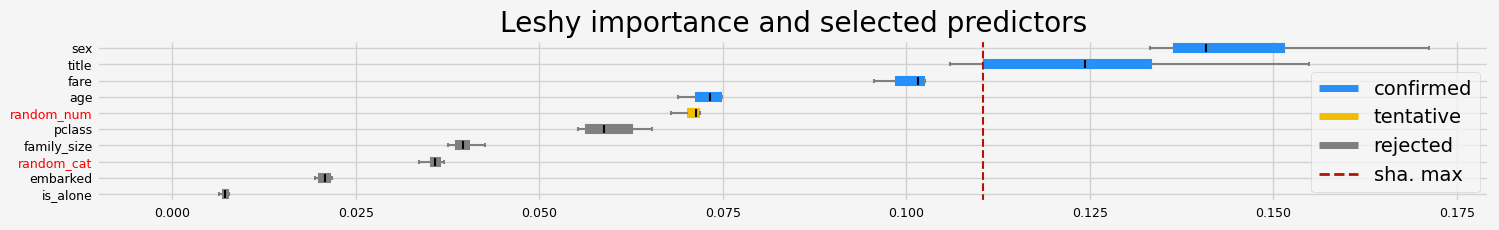
==================== Leshy - testing: CatBoostClassifier for var.imp: shap ====================
Leshy finished running using shap var. imp.
Iteration: 1 / 10
Confirmed: 7
Tentative: 0
Rejected: 3
All relevant predictors selected in 00:00:05.29
['pclass' 'sex' 'embarked' 'title' 'age' 'family_size' 'fare']
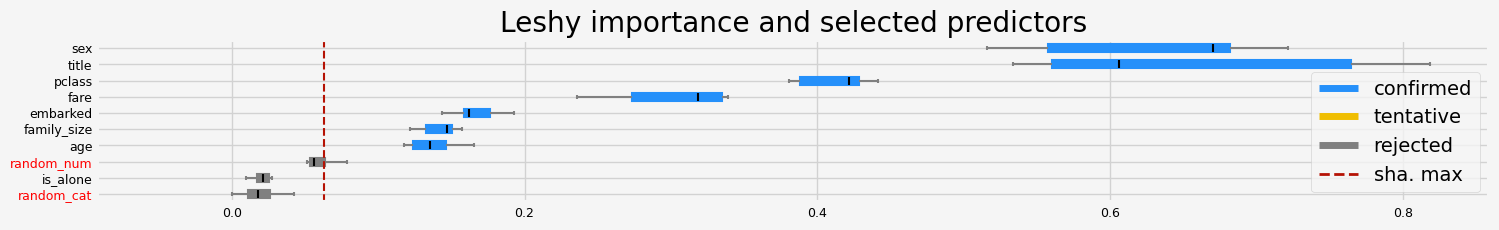
==================== Leshy - testing: CatBoostClassifier for var.imp: fastshap ====================
Leshy finished running using native var. imp.
Iteration: 1 / 10
Confirmed: 6
Tentative: 1
Rejected: 3
All relevant predictors selected in 00:00:05.04
['pclass' 'sex' 'embarked' 'title' 'family_size' 'fare']
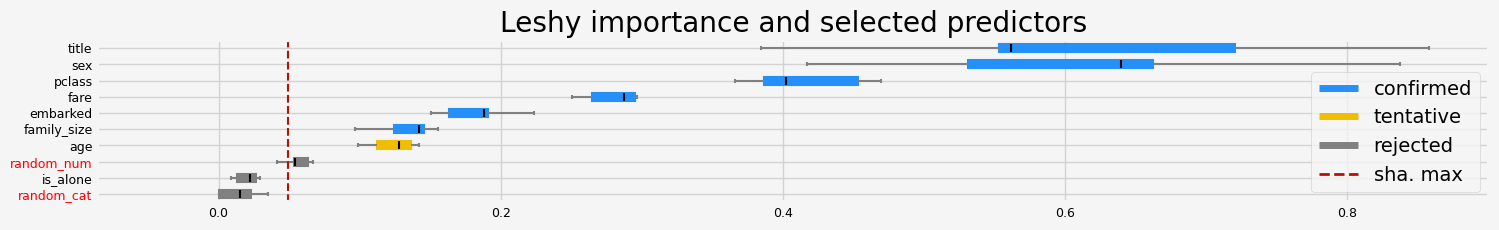
==================== Leshy - testing: CatBoostClassifier for var.imp: pimp ====================
Leshy finished running using pimp var. imp.
Iteration: 1 / 10
Confirmed: 4
Tentative: 2
Rejected: 4
All relevant predictors selected in 00:00:07.85
['pclass' 'sex' 'title' 'family_size']
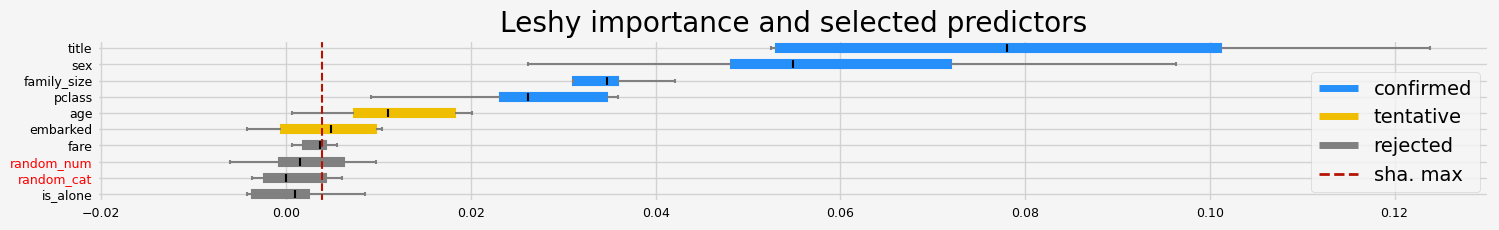
==================== Leshy - testing: CatBoostClassifier for var.imp: native ====================
Leshy finished running using native var. imp.
Iteration: 1 / 10
Confirmed: 5
Tentative: 2
Rejected: 3
All relevant predictors selected in 00:00:04.33
['pclass' 'sex' 'title' 'age' 'fare']
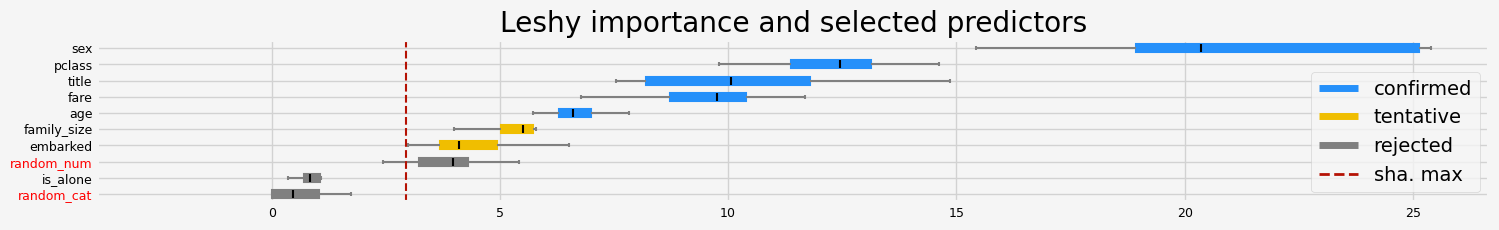
==================== Leshy - testing: LGBMClassifier for var.imp: shap ====================
Leshy finished running using shap var. imp.
Iteration: 1 / 10
Confirmed: 4
Tentative: 0
Rejected: 6
All relevant predictors selected in 00:00:03.22
['pclass' 'sex' 'age' 'fare']

==================== Leshy - testing: LGBMClassifier for var.imp: fastshap ====================
Leshy finished running using native var. imp.
Iteration: 1 / 10
Confirmed: 4
Tentative: 1
Rejected: 5
All relevant predictors selected in 00:00:03.48
['pclass' 'sex' 'age' 'fare']

==================== Leshy - testing: LGBMClassifier for var.imp: pimp ====================
Leshy finished running using pimp var. imp.
Iteration: 1 / 10
Confirmed: 4
Tentative: 1
Rejected: 5
All relevant predictors selected in 00:00:07.80
['pclass' 'sex' 'age' 'family_size']

==================== Leshy - testing: LGBMClassifier for var.imp: native ====================
Leshy finished running using native var. imp.
Iteration: 1 / 10
Confirmed: 1
Tentative: 1
Rejected: 8
All relevant predictors selected in 00:00:01.78
['fare']

==================== Leshy - testing: LGBMClassifier for var.imp: shap ====================
Leshy finished running using shap var. imp.
Iteration: 1 / 10
Confirmed: 5
Tentative: 2
Rejected: 3
All relevant predictors selected in 00:00:02.35
['pclass' 'sex' 'embarked' 'title' 'fare']
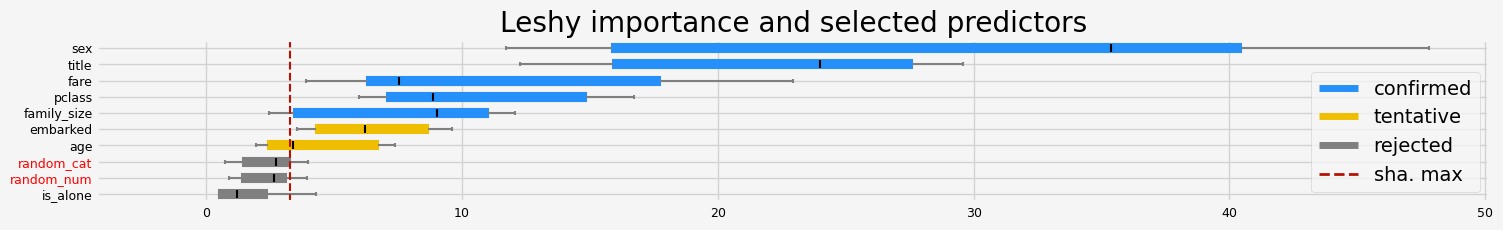
==================== Leshy - testing: LGBMClassifier for var.imp: fastshap ====================
Leshy finished running using native var. imp.
Iteration: 1 / 10
Confirmed: 6
Tentative: 1
Rejected: 3
All relevant predictors selected in 00:00:02.30
['pclass' 'sex' 'embarked' 'title' 'family_size' 'fare']
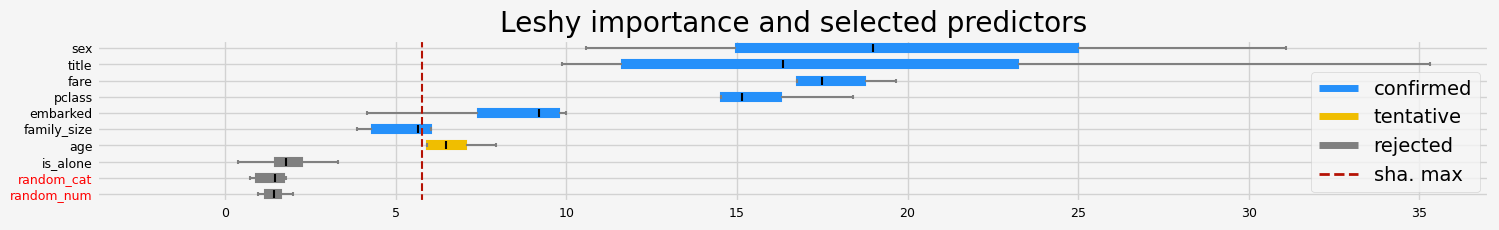
==================== Leshy - testing: LGBMClassifier for var.imp: pimp ====================
Leshy finished running using pimp var. imp.
Iteration: 1 / 10
Confirmed: 4
Tentative: 4
Rejected: 2
All relevant predictors selected in 00:00:06.88
['pclass' 'sex' 'title' 'family_size']
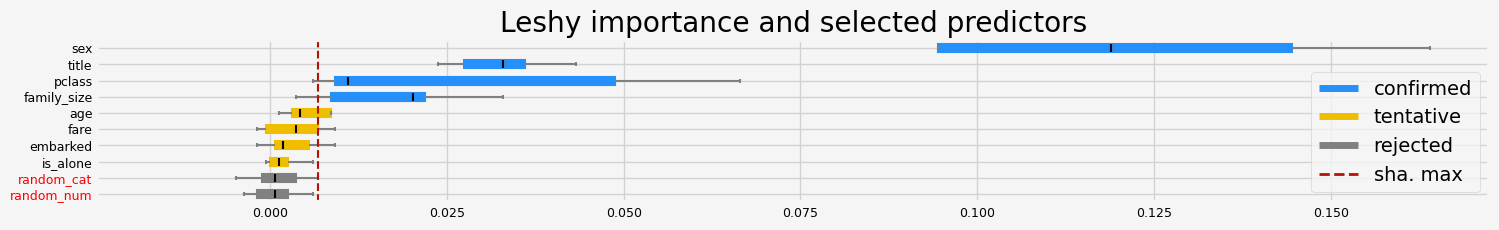
==================== Leshy - testing: LGBMClassifier for var.imp: native ====================
Leshy finished running using native var. imp.
Iteration: 1 / 10
Confirmed: 0
Tentative: 1
Rejected: 9
All relevant predictors selected in 00:00:01.60
[]
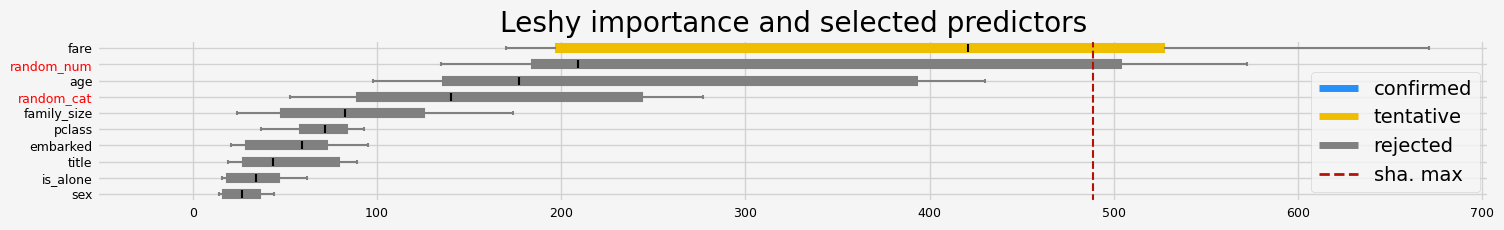
==================== Leshy - testing: XGBClassifier for var.imp: shap ====================
Leshy finished running using shap var. imp.
Iteration: 1 / 10
Confirmed: 4
Tentative: 2
Rejected: 4
All relevant predictors selected in 00:00:04.02
['pclass' 'sex' 'age' 'fare']
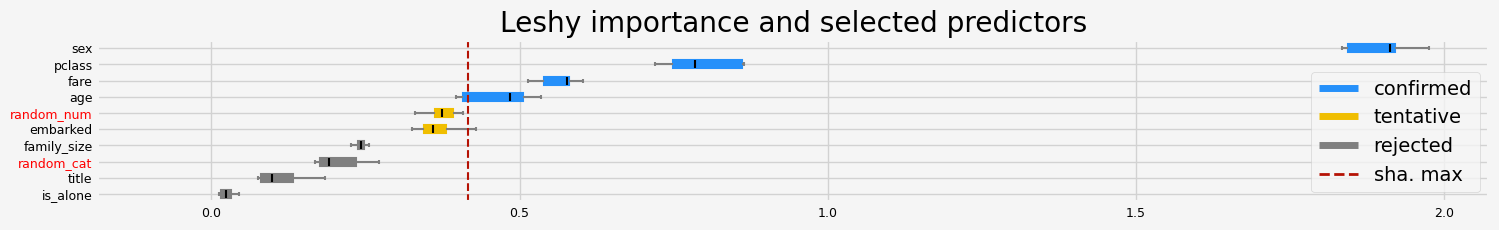
==================== Leshy - testing: XGBClassifier for var.imp: fastshap ====================
---------------------------------------------------------------------------
Exception Traceback (most recent call last)
Cell In[23], line 19
17 cat_f = titanic.categorical
18 # running the ARFS methods using different models
---> 19 compare_varimp(feat_selector, models, X, y, sample_weight=None)
File ~/Documents/arfs/src/arfs/benchmark.py:142, in compare_varimp(feat_selector, models, X, y, sample_weight)
140 feat_selector.estimator = mod_clone
141 # fit the feature selector
--> 142 feat_selector.fit(X=X, y=y, sample_weight=sample_weight)
143 # print the results
144 print(feat_selector.selected_features_)
File ~/Documents/arfs/src/arfs/feature_selection/allrelevant.py:329, in Leshy.fit(self, X, y, sample_weight)
326 raise TypeError("X is not a dataframe")
328 self.imp_real_hist = np.empty((0, X.shape[1]), float)
--> 329 self._fit(X, y, sample_weight=sample_weight)
330 self.selected_features_ = self.feature_names_in_[self.support_]
331 self.not_selected_features_ = self.feature_names_in_[~self.support_]
File ~/Documents/arfs/src/arfs/feature_selection/allrelevant.py:468, in Leshy._fit(self, X_raw, y, sample_weight)
465 if self.n_estimators != "auto":
466 self.estimator.set_params(n_estimators=self.n_estimators)
--> 468 dec_reg, sha_max_history, imp_history, imp_sha_max = self.select_features(
469 X=X, y=y, sample_weight=sample_weight
470 )
471 confirmed, tentative = _get_confirmed_and_tentative(dec_reg)
472 tentative = _select_tentative(tentative, imp_history, sha_max_history)
File ~/Documents/arfs/src/arfs/feature_selection/allrelevant.py:939, in Leshy.select_features(self, X, y, sample_weight)
931 self._update_tree_num(dec_reg)
932 self._update_estimator()
933 (
934 dec_reg,
935 sha_max_history,
936 imp_history,
937 hit_reg,
938 imp_sha_max,
--> 939 ) = self._run_iteration(
940 X,
941 y,
942 sample_weight,
943 dec_reg,
944 sha_max_history,
945 imp_history,
946 hit_reg,
947 _iter,
948 )
949 _iter += 1
950 pbar.update(1)
File ~/Documents/arfs/src/arfs/feature_selection/allrelevant.py:890, in Leshy._run_iteration(self, X, y, sample_weight, dec_reg, sha_max_history, imp_history, hit_reg, _iter)
841 def _run_iteration(
842 self, X, y, sample_weight, dec_reg, sha_max_history, imp_history, hit_reg, _iter
843 ):
844 """
845 Run an iteration of the Gradient Boosting algorithm.
846
(...)
888 The maximum shadow importance value for this iteration.
889 """
--> 890 cur_imp = self._add_shadows_get_imps(X, y, sample_weight, dec_reg)
891 imp_sha_max = np.percentile(cur_imp[1], self.perc)
892 sha_max_history.append(imp_sha_max)
File ~/Documents/arfs/src/arfs/feature_selection/allrelevant.py:557, in Leshy._add_shadows_get_imps(self, X, y, sample_weight, dec_reg)
553 imp = _get_shap_imp(
554 self.estimator, pd.concat([x_cur, x_sha], axis=1), y, sample_weight
555 )
556 elif self.importance == "fastshap":
--> 557 imp = _get_shap_imp_fast(
558 self.estimator, pd.concat([x_cur, x_sha], axis=1), y, sample_weight
559 )
560 elif self.importance == "pimp":
561 imp = _get_perm_imp(
562 self.estimator, pd.concat([x_cur, x_sha], axis=1), y, sample_weight
563 )
File ~/Documents/arfs/src/arfs/feature_selection/allrelevant.py:1301, in _get_shap_imp_fast(estimator, X, y, sample_weight, cat_feature)
1292 model, X_tt, y_tt, w_tt = _split_fit_estimator(
1293 estimator, X, y, sample_weight=sample_weight, cat_feature=cat_feature
1294 )
1295 explainer = FastTreeExplainer(
1296 model,
1297 algorithm="auto",
1298 shortcut=False,
1299 feature_perturbation="tree_path_dependent",
1300 )
-> 1301 shap_matrix = explainer.shap_values(X_tt)
1302 # multiclass returns a list
1303 # for binary and for some models, shap is still returning a list
1304 if is_classifier(estimator):
File ~/mambaforge-pypy3/envs/arfs/lib/python3.10/site-packages/fasttreeshap/explainers/_tree.py:481, in Tree.shap_values(self, X, y, tree_limit, approximate, check_additivity, from_call)
479 out = self._get_shap_output(phi, flat_output)
480 if check_additivity and self.model.model_output == "raw":
--> 481 self.assert_additivity(out, self.model.predict(X))
483 return out
File ~/mambaforge-pypy3/envs/arfs/lib/python3.10/site-packages/fasttreeshap/explainers/_tree.py:641, in Tree.assert_additivity(self, phi, model_output)
639 check_sum(self.expected_value[i] + phi[i].sum(-1), model_output[:,i])
640 else:
--> 641 check_sum(self.expected_value + phi.sum(-1), model_output)
File ~/mambaforge-pypy3/envs/arfs/lib/python3.10/site-packages/fasttreeshap/explainers/_tree.py:635, in Tree.assert_additivity.<locals>.check_sum(sum_val, model_output)
631 err_msg += " Consider retrying with the feature_perturbation='interventional' option."
632 err_msg += " This check failed because for one of the samples the sum of the SHAP values" \
633 " was %f, while the model output was %f. If this difference is acceptable" \
634 " you can set check_additivity=False to disable this check." % (sum_val[ind], model_output[ind])
--> 635 raise Exception(err_msg)
Exception: Additivity check failed in TreeExplainer! Please ensure the data matrix you passed to the explainer is the same shape that the model was trained on. If your data shape is correct then please report this on GitHub. Consider retrying with the feature_perturbation='interventional' option. This check failed because for one of the samples the sum of the SHAP values was -4.916668, while the model output was -5.572705. If this difference is acceptable you can set check_additivity=False to disable this check.
[24]:
from sklearn.datasets import make_classification
from xgboost import XGBClassifier
from fasttreeshap import TreeExplainer as FastTreeExplainer
X, y = make_classification(
n_samples=1000, n_features=10, n_informative=8, random_state=8
)
model = XGBClassifier()
model.fit(X, y)
explainer = FastTreeExplainer(
model, algorithm="auto", shortcut=False, feature_perturbation="tree_path_dependent"
)
shap_matrix = explainer.shap_values(X)
---------------------------------------------------------------------------
Exception Traceback (most recent call last)
Cell In[24], line 13
9 model.fit(X, y)
10 explainer = FastTreeExplainer(
11 model, algorithm="auto", shortcut=False, feature_perturbation="tree_path_dependent"
12 )
---> 13 shap_matrix = explainer.shap_values(X)
File ~/mambaforge-pypy3/envs/arfs/lib/python3.10/site-packages/fasttreeshap/explainers/_tree.py:481, in Tree.shap_values(self, X, y, tree_limit, approximate, check_additivity, from_call)
479 out = self._get_shap_output(phi, flat_output)
480 if check_additivity and self.model.model_output == "raw":
--> 481 self.assert_additivity(out, self.model.predict(X))
483 return out
File ~/mambaforge-pypy3/envs/arfs/lib/python3.10/site-packages/fasttreeshap/explainers/_tree.py:641, in Tree.assert_additivity(self, phi, model_output)
639 check_sum(self.expected_value[i] + phi[i].sum(-1), model_output[:,i])
640 else:
--> 641 check_sum(self.expected_value + phi.sum(-1), model_output)
File ~/mambaforge-pypy3/envs/arfs/lib/python3.10/site-packages/fasttreeshap/explainers/_tree.py:635, in Tree.assert_additivity.<locals>.check_sum(sum_val, model_output)
631 err_msg += " Consider retrying with the feature_perturbation='interventional' option."
632 err_msg += " This check failed because for one of the samples the sum of the SHAP values" \
633 " was %f, while the model output was %f. If this difference is acceptable" \
634 " you can set check_additivity=False to disable this check." % (sum_val[ind], model_output[ind])
--> 635 raise Exception(err_msg)
Exception: Additivity check failed in TreeExplainer! Please ensure the data matrix you passed to the explainer is the same shape that the model was trained on. If your data shape is correct then please report this on GitHub. Consider retrying with the feature_perturbation='interventional' option. This check failed because for one of the samples the sum of the SHAP values was 4.292476, while the model output was 4.248128. If this difference is acceptable you can set check_additivity=False to disable this check.
the fasttreeshap implementation doesn’t work correctly with xgboost, I created an issue
Testing GrootCV#
[25]:
# Testing the changes with rnd cat. and num. predictors added to the set of genuine predictors
def testing_estimators(X, y, sample_weight=None, objective="binary"):
feat_selector = arfsgroot.GrootCV(
objective=objective, cutoff=1, n_folds=5, n_iter=5
)
feat_selector.fit(X, y, sample_weight)
print(feat_selector.get_feature_names_out())
fig = feat_selector.plot_importance(n_feat_per_inch=5)
# highlight synthetic random variable
fig = highlight_tick(figure=fig, str_match="random")
plt.show()
gc.enable()
del feat_selector
gc.collect()
if __name__ == "__main__":
# classification
titanic = load_data(name="Titanic")
X, y = titanic.data, titanic.target
y = y.astype(int)
cat_f = titanic.categorical
testing_estimators(X=X, y=y, objective="binary")
The default value of `parser` will change from `'liac-arff'` to `'auto'` in 1.4. You can set `parser='auto'` to silence this warning. Therefore, an `ImportError` will be raised from 1.4 if the dataset is dense and pandas is not installed. Note that the pandas parser may return different data types. See the Notes Section in fetch_openml's API doc for details.
['pclass' 'sex' 'age' 'fare']
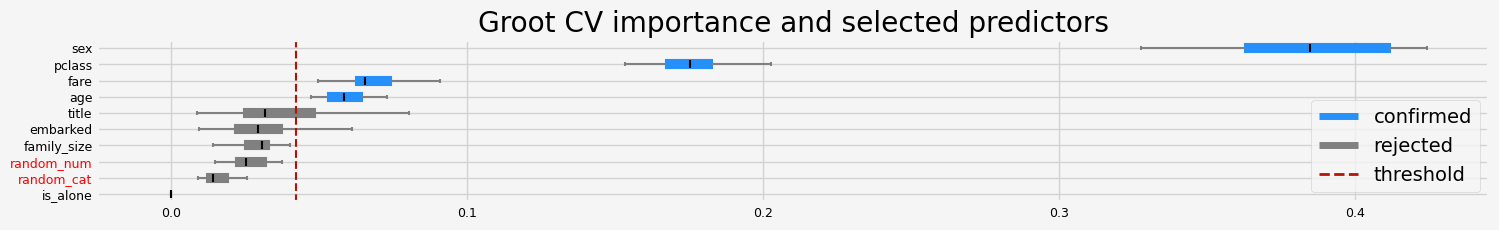
This confirms that the native (gini/gain) feature importance are biased and not the best to assess the real feature importance.
Testing BoostAGroota#
[26]:
models = [
RandomForestClassifier(n_jobs=4, oob_score=True),
CatBoostClassifier(random_state=42, verbose=0),
LGBMClassifier(random_state=42, verbose=-1),
LightForestClassifier(n_feat=X.shape[1]),
XGBClassifier(random_state=42, verbosity=0),
]
feat_selector = arfsgroot.BoostAGroota(
estimator=model, cutoff=1.25, iters=10, max_rounds=10, delta=0.1
)
if __name__ == "__main__":
# classification
titanic = load_data(name="Titanic")
X, y = titanic.data, titanic.target
y = y.astype(int)
cat_f = titanic.categorical
# running the ARFS methods using different models
compare_varimp(feat_selector, models, X, y, sample_weight=None)
The default value of `parser` will change from `'liac-arff'` to `'auto'` in 1.4. You can set `parser='auto'` to silence this warning. Therefore, an `ImportError` will be raised from 1.4 if the dataset is dense and pandas is not installed. Note that the pandas parser may return different data types. See the Notes Section in fetch_openml's API doc for details.
==================== BoostAGroota - testing: RandomForestClassifier for var.imp: shap ====================
['pclass' 'sex' 'embarked' 'title' 'age' 'family_size' 'fare']
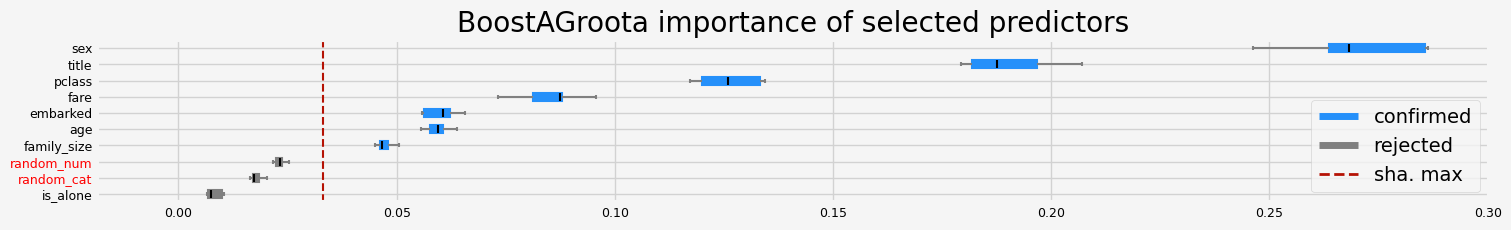
==================== BoostAGroota - testing: RandomForestClassifier for var.imp: fastshap ====================
['pclass' 'sex' 'embarked' 'title' 'age' 'family_size' 'fare']
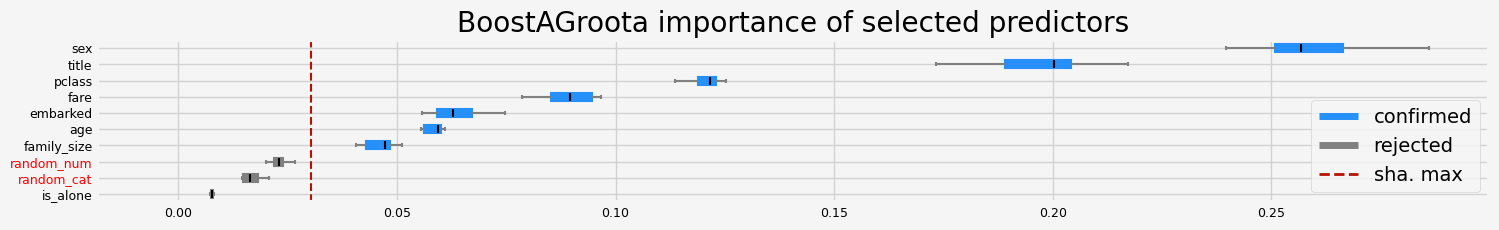
==================== BoostAGroota - testing: RandomForestClassifier for var.imp: pimp ====================
['pclass' 'sex' 'title' 'age' 'family_size']
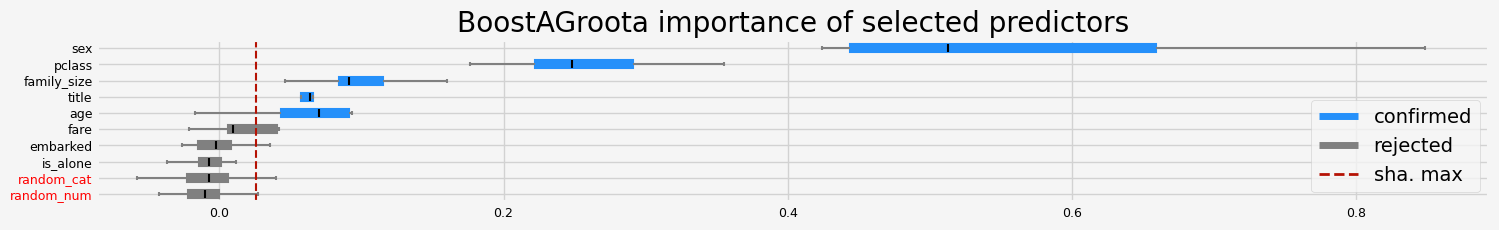
==================== BoostAGroota - testing: RandomForestClassifier for var.imp: native ====================
['sex' 'title' 'age' 'fare' 'random_num']
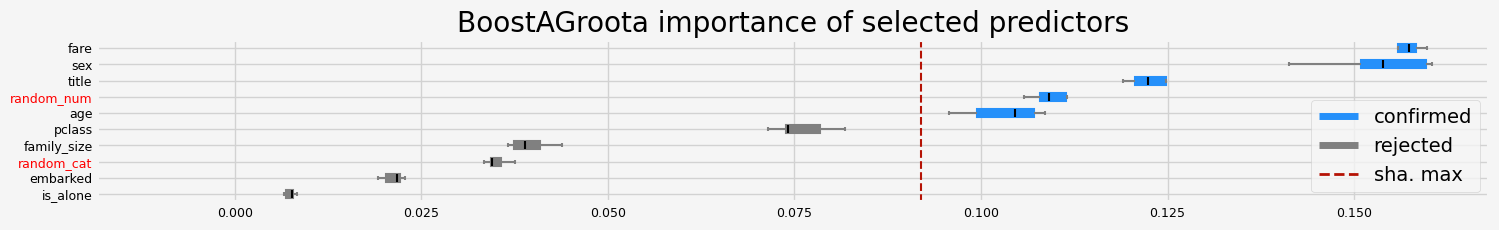
==================== BoostAGroota - testing: CatBoostClassifier for var.imp: shap ====================
['pclass' 'sex' 'embarked' 'title' 'age' 'family_size' 'fare']
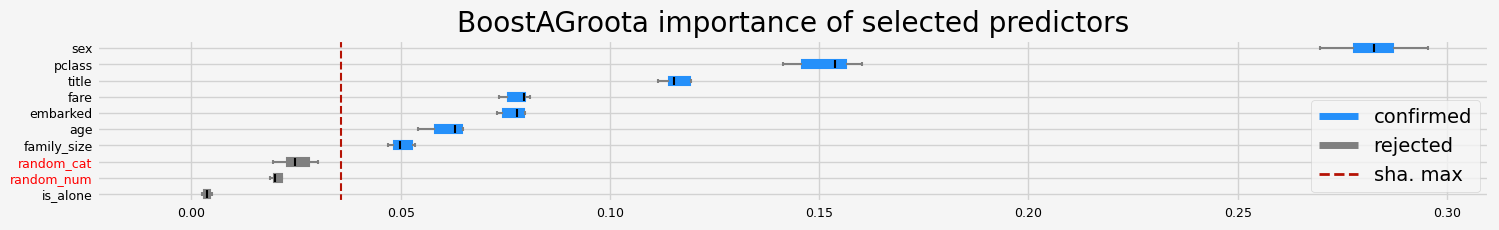
==================== BoostAGroota - testing: CatBoostClassifier for var.imp: fastshap ====================
['pclass' 'sex' 'embarked' 'title' 'age' 'family_size' 'fare']
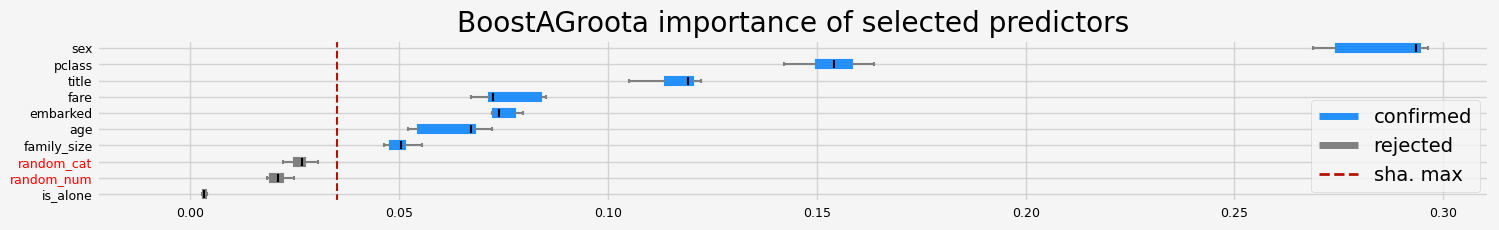
==================== BoostAGroota - testing: CatBoostClassifier for var.imp: pimp ====================
['pclass' 'sex' 'title' 'age' 'family_size']
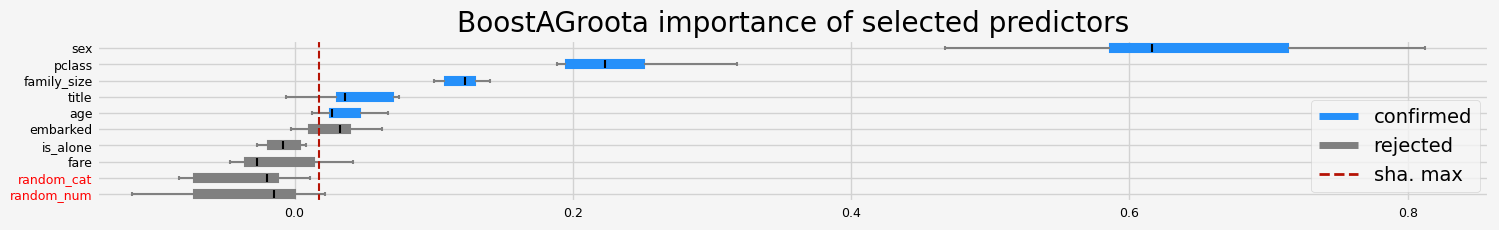
==================== BoostAGroota - testing: CatBoostClassifier for var.imp: native ====================
['pclass' 'sex' 'title' 'age' 'fare' 'random_num']
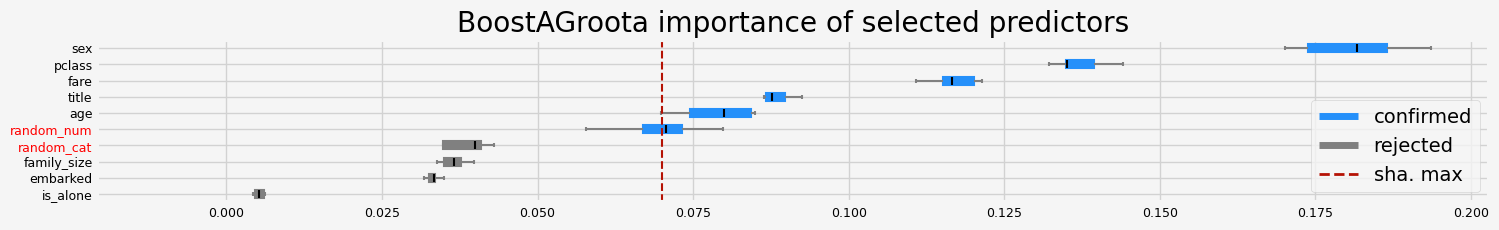
==================== BoostAGroota - testing: LGBMClassifier for var.imp: shap ====================
['pclass' 'sex' 'age' 'fare']
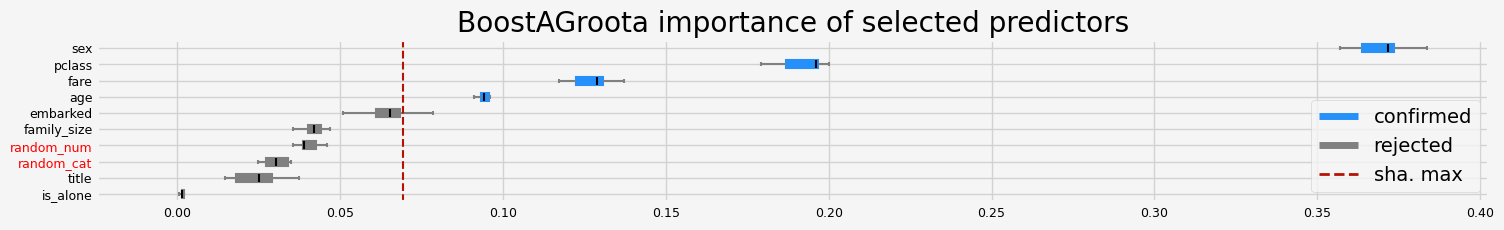
==================== BoostAGroota - testing: LGBMClassifier for var.imp: fastshap ====================
['pclass' 'sex' 'age' 'fare']
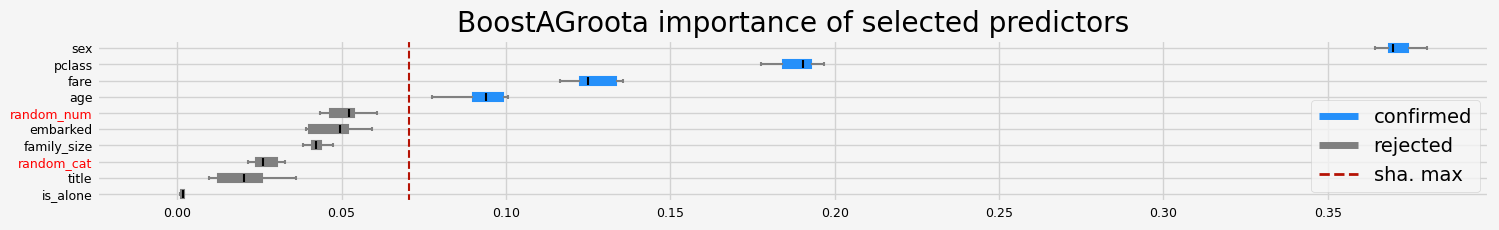
==================== BoostAGroota - testing: LGBMClassifier for var.imp: pimp ====================
['pclass' 'sex' 'age' 'family_size' 'fare']
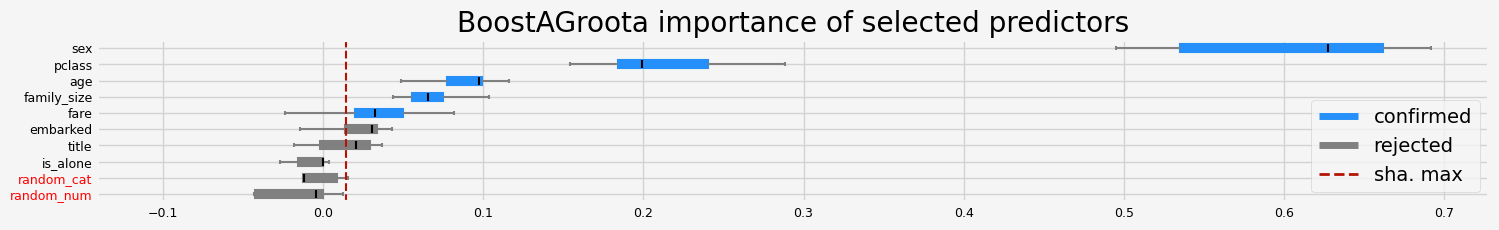
==================== BoostAGroota - testing: LGBMClassifier for var.imp: native ====================
['fare' 'random_num']
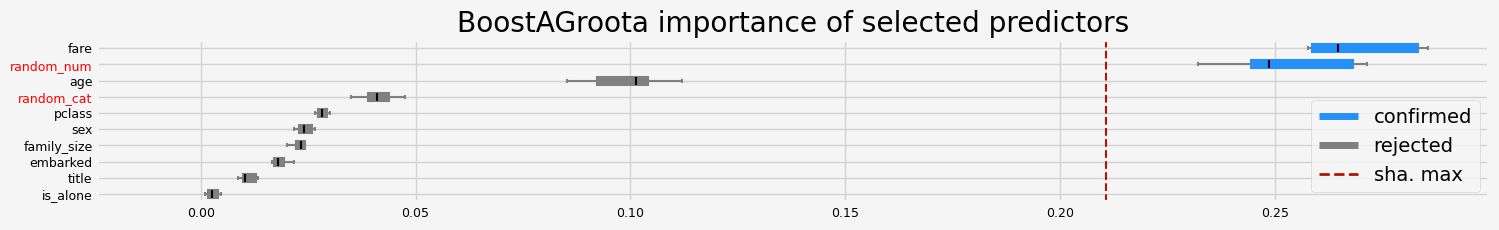
==================== BoostAGroota - testing: LGBMClassifier for var.imp: shap ====================
['pclass' 'sex' 'title' 'family_size']
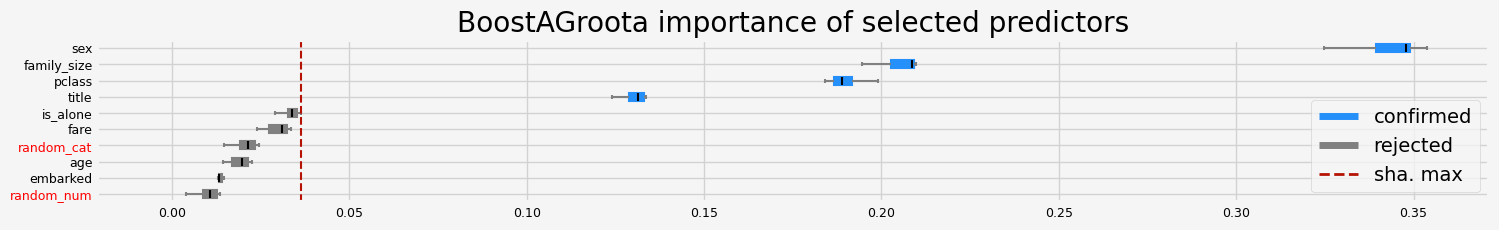
==================== BoostAGroota - testing: LGBMClassifier for var.imp: fastshap ====================
['pclass' 'sex' 'title' 'fare']
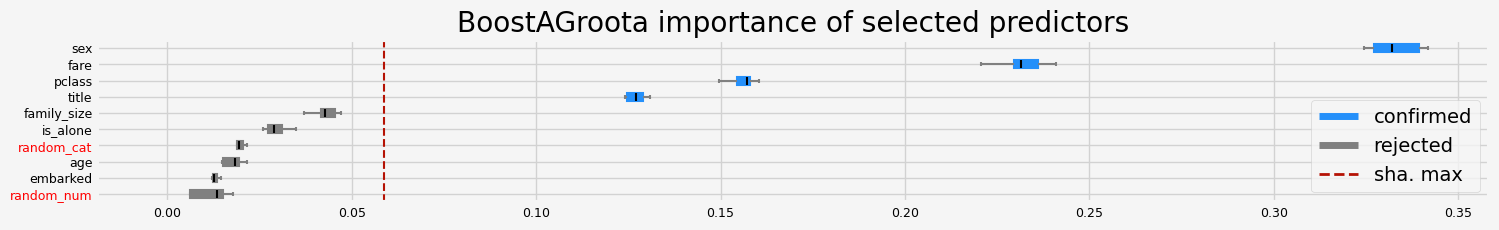
==================== BoostAGroota - testing: LGBMClassifier for var.imp: pimp ====================
['pclass' 'sex' 'title' 'fare']
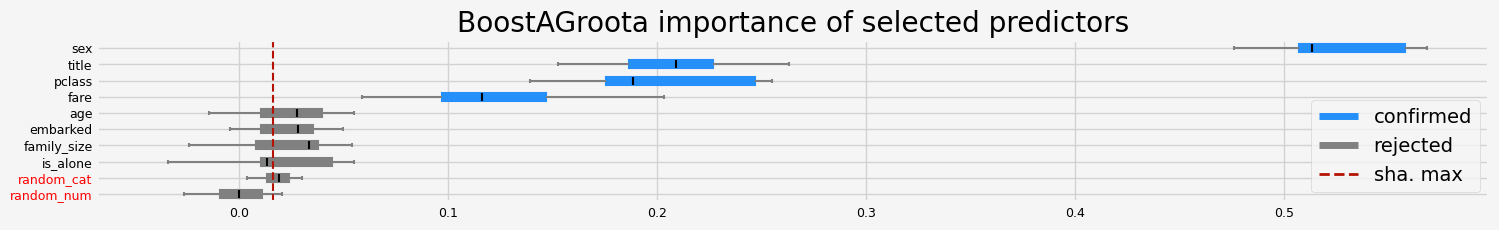
==================== BoostAGroota - testing: LGBMClassifier for var.imp: native ====================
[]
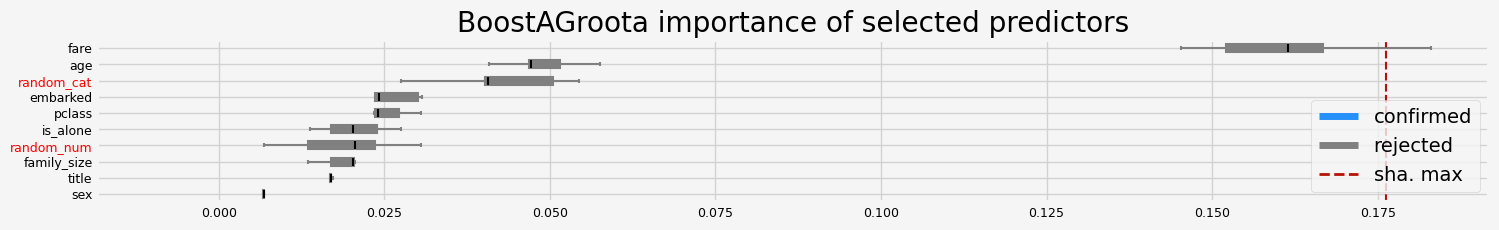
==================== BoostAGroota - testing: XGBClassifier for var.imp: shap ====================
['pclass' 'sex' 'age' 'fare' 'random_num']
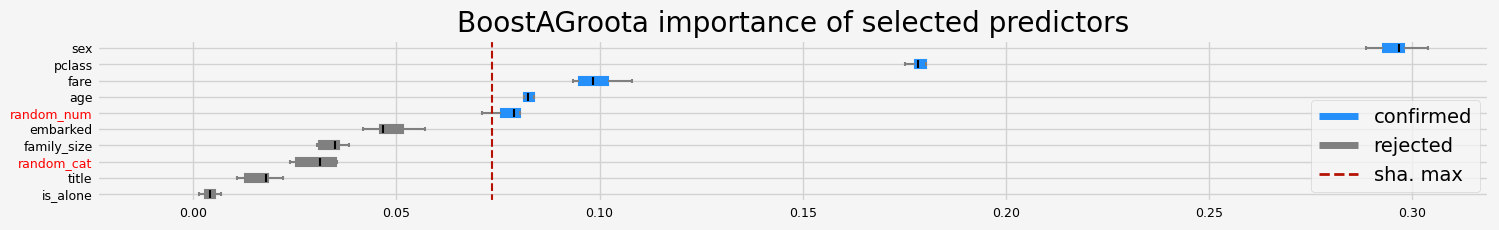
==================== BoostAGroota - testing: XGBClassifier for var.imp: fastshap ====================
['pclass' 'sex' 'age' 'fare' 'random_num']
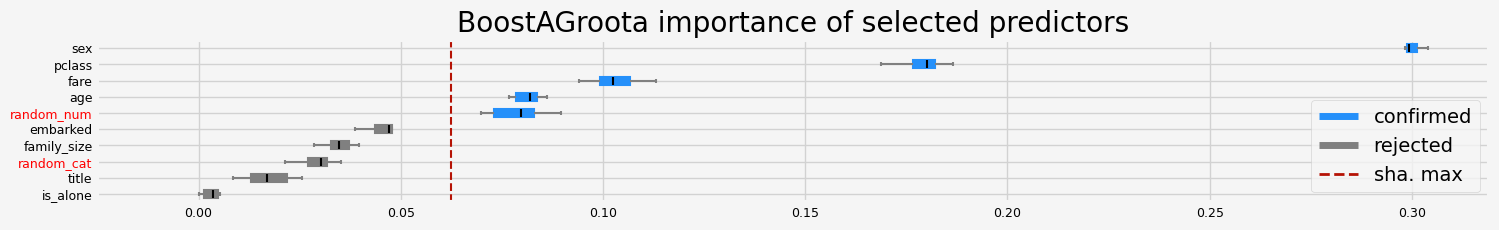
==================== BoostAGroota - testing: XGBClassifier for var.imp: pimp ====================
['pclass' 'sex' 'age' 'family_size' 'fare']
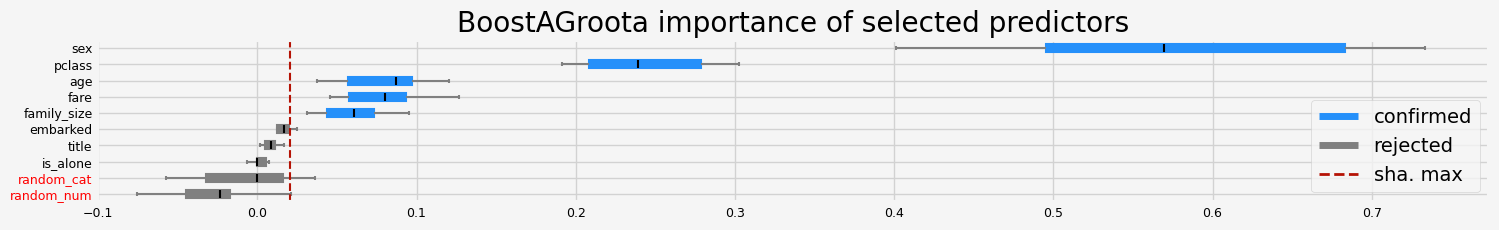
==================== BoostAGroota - testing: XGBClassifier for var.imp: native ====================
['pclass' 'sex' 'embarked' 'title' 'age' 'family_size' 'fare']

comparing to BorutaShap#
[27]:
from BorutaShap import BorutaShap
from arfs.preprocessing import OrdinalEncoderPandas
model = LGBMClassifier(random_state=42, verbose=-1, n_estimators=10)
X_encoded = OrdinalEncoderPandas().fit_transform(X=X)
Feature_Selector = BorutaShap(
model=model, importance_measure="shap", classification=True
)
Feature_Selector.fit(X=X_encoded, y=y, n_trials=100, random_state=0)
# Returns Boxplot of features
Feature_Selector.plot(X_size=12, figsize=(8, 6), y_scale="log", which_features="all")
---------------------------------------------------------------------------
ImportError Traceback (most recent call last)
Cell In[27], line 1
----> 1 from BorutaShap import BorutaShap
2 from arfs.preprocessing import OrdinalEncoderPandas
4 model = LGBMClassifier(random_state=42, verbose=-1, n_estimators=10)
File ~/mambaforge-pypy3/envs/arfs/lib/python3.10/site-packages/BorutaShap.py:2
1 from sklearn.ensemble import RandomForestClassifier, RandomForestRegressor, IsolationForest
----> 2 from sklearn.datasets import load_breast_cancer, load_boston
3 from statsmodels.stats.multitest import multipletests
4 from sklearn.model_selection import train_test_split
File ~/mambaforge-pypy3/envs/arfs/lib/python3.10/site-packages/sklearn/datasets/__init__.py:157, in __getattr__(name)
108 if name == "load_boston":
109 msg = textwrap.dedent("""
110 `load_boston` has been removed from scikit-learn since version 1.2.
111
(...)
155 <https://www.researchgate.net/publication/4974606_Hedonic_housing_prices_and_the_demand_for_clean_air>
156 """)
--> 157 raise ImportError(msg)
158 try:
159 return globals()[name]
ImportError:
`load_boston` has been removed from scikit-learn since version 1.2.
The Boston housing prices dataset has an ethical problem: as
investigated in [1], the authors of this dataset engineered a
non-invertible variable "B" assuming that racial self-segregation had a
positive impact on house prices [2]. Furthermore the goal of the
research that led to the creation of this dataset was to study the
impact of air quality but it did not give adequate demonstration of the
validity of this assumption.
The scikit-learn maintainers therefore strongly discourage the use of
this dataset unless the purpose of the code is to study and educate
about ethical issues in data science and machine learning.
In this special case, you can fetch the dataset from the original
source::
import pandas as pd
import numpy as np
data_url = "http://lib.stat.cmu.edu/datasets/boston"
raw_df = pd.read_csv(data_url, sep="\s+", skiprows=22, header=None)
data = np.hstack([raw_df.values[::2, :], raw_df.values[1::2, :2]])
target = raw_df.values[1::2, 2]
Alternative datasets include the California housing dataset and the
Ames housing dataset. You can load the datasets as follows::
from sklearn.datasets import fetch_california_housing
housing = fetch_california_housing()
for the California housing dataset and::
from sklearn.datasets import fetch_openml
housing = fetch_openml(name="house_prices", as_frame=True)
for the Ames housing dataset.
[1] M Carlisle.
"Racist data destruction?"
<https://medium.com/@docintangible/racist-data-destruction-113e3eff54a8>
[2] Harrison Jr, David, and Daniel L. Rubinfeld.
"Hedonic housing prices and the demand for clean air."
Journal of environmental economics and management 5.1 (1978): 81-102.
<https://www.researchgate.net/publication/4974606_Hedonic_housing_prices_and_the_demand_for_clean_air>
[28]:
# Leshy
from arfs.preprocessing import OrdinalEncoderPandas
model = LGBMClassifier(random_state=42, verbose=-1, n_estimators=10)
X_encoded = OrdinalEncoderPandas().fit_transform(X=X)
feat_selector = arfsgroot.Leshy(
model, n_estimators=10, verbose=1, max_iter=100, random_state=42, importance="shap"
)
feat_selector.fit(X_encoded, y, sample_weight=None)
print(feat_selector.get_feature_names_out())
fig = feat_selector.plot_importance(n_feat_per_inch=5)
# highlight synthetic random variable
fig = highlight_tick(figure=fig, str_match="random")
fig = highlight_tick(figure=fig, str_match="genuine", color="green")
plt.show()
Leshy finished running using shap var. imp.
Iteration: 1 / 100
Confirmed: 5
Tentative: 0
Rejected: 5
All relevant predictors selected in 00:00:05.05
['pclass' 'sex' 'embarked' 'age' 'fare']